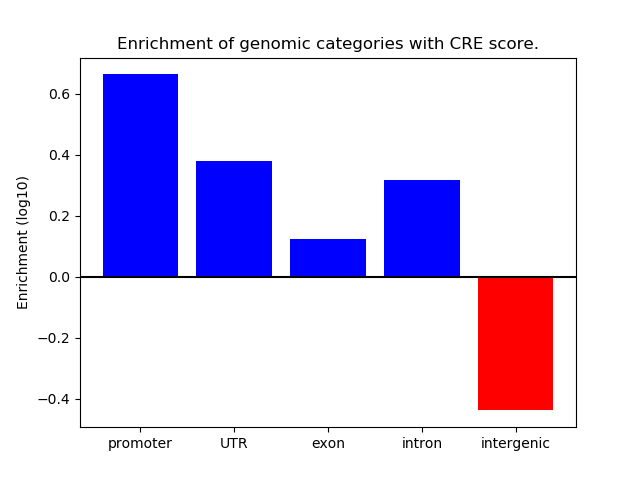
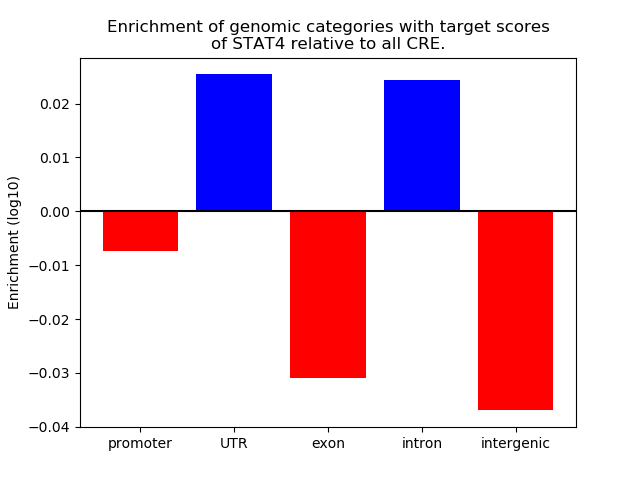
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
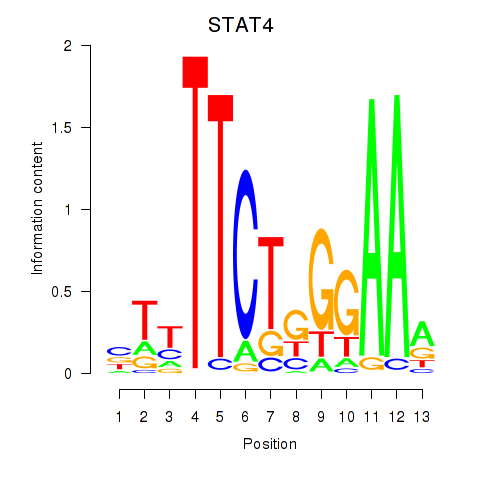
Results for STAT4
Z-value: 1.20
Transcription factors associated with STAT4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT4
|
ENSG00000138378.13 | signal transducer and activator of transcription 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_192013340_192013491 | STAT4 | 2282 | 0.339131 | 0.66 | 5.3e-02 | Click! |
| chr2_192016485_192016636 | STAT4 | 238 | 0.941695 | 0.56 | 1.2e-01 | Click! |
| chr2_192056884_192057039 | STAT4 | 40639 | 0.156524 | -0.49 | 1.8e-01 | Click! |
| chr2_192012454_192012731 | STAT4 | 3105 | 0.286995 | 0.48 | 1.9e-01 | Click! |
| chr2_192037184_192037335 | STAT4 | 20937 | 0.205608 | -0.45 | 2.2e-01 | Click! |
Activity of the STAT4 motif across conditions
Conditions sorted by the z-value of the STAT4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_136013025_136013297 | 0.45 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
6657 |
0.15 |
| chr14_75038764_75038974 | 0.41 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
39906 |
0.13 |
| chr5_156621341_156621645 | 0.39 |
ITK |
IL2-inducible T-cell kinase |
13656 |
0.12 |
| chr1_66518298_66518449 | 0.37 |
RP11-397C12.1 |
|
1972 |
0.41 |
| chr7_24940112_24940263 | 0.35 |
ENSG00000238976 |
. |
244 |
0.94 |
| chr1_90097018_90097733 | 0.34 |
RP5-1007M22.2 |
|
375 |
0.77 |
| chr1_112059507_112059886 | 0.32 |
ADORA3 |
adenosine A3 receptor |
13314 |
0.12 |
| chr12_12647697_12647848 | 0.30 |
DUSP16 |
dual specificity phosphatase 16 |
26287 |
0.21 |
| chr7_3102888_3103135 | 0.30 |
CARD11 |
caspase recruitment domain family, member 11 |
19432 |
0.23 |
| chr4_109015585_109015736 | 0.29 |
LEF1 |
lymphoid enhancer-binding factor 1 |
71797 |
0.1 |
| chr7_144516745_144516896 | 0.29 |
TPK1 |
thiamin pyrophosphokinase 1 |
16326 |
0.24 |
| chr13_43149418_43149818 | 0.29 |
TNFSF11 |
tumor necrosis factor (ligand) superfamily, member 11 |
1258 |
0.64 |
| chr18_35289879_35290030 | 0.28 |
ENSG00000266530 |
. |
52856 |
0.18 |
| chr4_122111518_122111869 | 0.28 |
ENSG00000252183 |
. |
2365 |
0.34 |
| chr5_156953479_156953635 | 0.28 |
ADAM19 |
ADAM metallopeptidase domain 19 |
23727 |
0.15 |
| chr6_159228545_159229058 | 0.28 |
EZR-AS1 |
EZR antisense RNA 1 |
10242 |
0.17 |
| chr10_115610846_115611146 | 0.27 |
DCLRE1A |
DNA cross-link repair 1A |
2863 |
0.22 |
| chr1_112192925_112193076 | 0.27 |
ENSG00000201028 |
. |
159 |
0.95 |
| chr6_25036886_25037064 | 0.26 |
RP3-425P12.5 |
|
5092 |
0.19 |
| chr3_31504876_31505139 | 0.26 |
ENSG00000238727 |
. |
67578 |
0.12 |
| chr7_75248927_75249078 | 0.26 |
HIP1 |
huntingtin interacting protein 1 |
7841 |
0.2 |
| chr7_21391785_21392069 | 0.26 |
ENSG00000195024 |
. |
29054 |
0.2 |
| chr14_63670641_63670908 | 0.26 |
RHOJ |
ras homolog family member J |
352 |
0.91 |
| chr15_60863549_60863700 | 0.26 |
RORA |
RAR-related orphan receptor A |
21116 |
0.19 |
| chr2_28836401_28836726 | 0.26 |
PLB1 |
phospholipase B1 |
11777 |
0.21 |
| chr18_22242340_22242491 | 0.25 |
HRH4 |
histamine receptor H4 |
201722 |
0.02 |
| chr11_126336020_126336230 | 0.25 |
KIRREL3 |
kin of IRRE like 3 (Drosophila) |
25672 |
0.18 |
| chr14_38540644_38540853 | 0.24 |
CTD-2058B24.2 |
|
19615 |
0.24 |
| chr1_167468557_167468835 | 0.24 |
CD247 |
CD247 molecule |
19079 |
0.18 |
| chr17_37937298_37937847 | 0.24 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
3094 |
0.19 |
| chr3_113250406_113250557 | 0.24 |
SIDT1 |
SID1 transmembrane family, member 1 |
662 |
0.69 |
| chr4_143634715_143634901 | 0.24 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
132635 |
0.06 |
| chr2_68948486_68948822 | 0.24 |
ARHGAP25 |
Rho GTPase activating protein 25 |
11020 |
0.25 |
| chr20_30392262_30392413 | 0.24 |
MYLK2 |
myosin light chain kinase 2 |
14774 |
0.14 |
| chr7_17068483_17068634 | 0.24 |
AGR3 |
anterior gradient 3 |
146947 |
0.04 |
| chr2_42352864_42353761 | 0.24 |
EML4 |
echinoderm microtubule associated protein like 4 |
43178 |
0.15 |
| chr13_99209848_99210037 | 0.23 |
STK24 |
serine/threonine kinase 24 |
19175 |
0.2 |
| chr10_21807546_21807797 | 0.23 |
SKIDA1 |
SKI/DACH domain containing 1 |
823 |
0.52 |
| chr3_17341812_17341972 | 0.23 |
TBC1D5 |
TBC1 domain family, member 5 |
106320 |
0.08 |
| chr3_196291691_196292106 | 0.23 |
WDR53 |
WD repeat domain 53 |
2283 |
0.21 |
| chr15_38976681_38977159 | 0.23 |
C15orf53 |
chromosome 15 open reading frame 53 |
11879 |
0.28 |
| chr4_186173181_186173380 | 0.23 |
SNX25 |
sorting nexin 25 |
42047 |
0.13 |
| chr21_34302222_34302373 | 0.23 |
AP000281.2 |
|
80053 |
0.07 |
| chr1_206942223_206942745 | 0.23 |
IL10 |
interleukin 10 |
3355 |
0.2 |
| chr3_53072289_53072451 | 0.23 |
SFMBT1 |
Scm-like with four mbt domains 1 |
6911 |
0.19 |
| chr20_57735088_57735478 | 0.23 |
ZNF831 |
zinc finger protein 831 |
30792 |
0.19 |
| chr2_55193973_55194124 | 0.23 |
RTN4 |
reticulon 4 |
43537 |
0.15 |
| chr1_197737430_197737581 | 0.23 |
DENND1B |
DENN/MADD domain containing 1B |
6817 |
0.23 |
| chr15_64454533_64454724 | 0.23 |
PPIB |
peptidylprolyl isomerase B (cyclophilin B) |
776 |
0.59 |
| chr19_47935849_47936000 | 0.22 |
MEIS3 |
Meis homeobox 3 |
13144 |
0.14 |
| chr2_204854148_204854486 | 0.22 |
ICOS |
inducible T-cell co-stimulator |
52814 |
0.17 |
| chr3_45983815_45984178 | 0.22 |
CXCR6 |
chemokine (C-X-C motif) receptor 6 |
968 |
0.5 |
| chr16_67018785_67019348 | 0.22 |
CES4A |
carboxylesterase 4A |
3426 |
0.14 |
| chr17_71588187_71588481 | 0.22 |
RP11-277J6.2 |
|
48831 |
0.13 |
| chr11_18640352_18640885 | 0.22 |
SPTY2D1 |
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
15422 |
0.14 |
| chr3_50609823_50610526 | 0.21 |
C3orf18 |
chromosome 3 open reading frame 18 |
1752 |
0.28 |
| chr14_22627506_22627657 | 0.21 |
ENSG00000238634 |
. |
16694 |
0.27 |
| chr16_87495374_87495580 | 0.21 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
28737 |
0.15 |
| chr14_50437257_50437517 | 0.21 |
C14orf182 |
chromosome 14 open reading frame 182 |
36851 |
0.12 |
| chr20_49168238_49168495 | 0.21 |
ENSG00000239742 |
. |
6169 |
0.15 |
| chr9_121284831_121284982 | 0.21 |
ENSG00000251847 |
. |
791157 |
0.0 |
| chr5_75622593_75622811 | 0.21 |
RP11-466P24.6 |
|
15415 |
0.26 |
| chr9_101854958_101855213 | 0.21 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
11235 |
0.23 |
| chr10_18069015_18069166 | 0.21 |
TMEM236 |
|
27863 |
0.17 |
| chr8_67452033_67452620 | 0.21 |
ENSG00000206949 |
. |
38780 |
0.13 |
| chr22_36558393_36558682 | 0.21 |
APOL3 |
apolipoprotein L, 3 |
1560 |
0.43 |
| chr11_118109252_118109403 | 0.20 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
13518 |
0.13 |
| chr19_11085110_11085263 | 0.20 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
9642 |
0.13 |
| chr10_69371869_69372293 | 0.20 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
35190 |
0.23 |
| chr5_131732657_131732808 | 0.20 |
C5orf56 |
chromosome 5 open reading frame 56 |
13596 |
0.12 |
| chr6_130505643_130505870 | 0.20 |
SAMD3 |
sterile alpha motif domain containing 3 |
29758 |
0.2 |
| chr10_496561_496809 | 0.20 |
RP11-490E15.2 |
|
13184 |
0.2 |
| chr20_33131898_33132049 | 0.20 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
2685 |
0.23 |
| chr8_2078833_2079052 | 0.20 |
MYOM2 |
myomesin 2 |
85758 |
0.1 |
| chr1_26031102_26031259 | 0.20 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
4913 |
0.2 |
| chr4_116134565_116134716 | 0.20 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
99608 |
0.09 |
| chr2_225790470_225790675 | 0.20 |
DOCK10 |
dedicator of cytokinesis 10 |
21210 |
0.27 |
| chr11_118123821_118123979 | 0.20 |
MPZL3 |
myelin protein zero-like 3 |
835 |
0.52 |
| chr7_50431752_50432472 | 0.19 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
64867 |
0.11 |
| chr1_43399996_43400147 | 0.19 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
3275 |
0.24 |
| chr8_59569855_59570006 | 0.19 |
NSMAF |
neutral sphingomyelinase (N-SMase) activation associated factor |
2163 |
0.35 |
| chr2_64978837_64979534 | 0.19 |
ENSG00000253082 |
. |
24053 |
0.19 |
| chr6_118978234_118978481 | 0.19 |
CEP85L |
centrosomal protein 85kDa-like |
4674 |
0.33 |
| chr5_75586536_75586687 | 0.19 |
RP11-466P24.6 |
|
20676 |
0.26 |
| chr13_74601733_74602123 | 0.19 |
KLF12 |
Kruppel-like factor 12 |
32742 |
0.25 |
| chr11_58407663_58407869 | 0.19 |
CNTF |
ciliary neurotrophic factor |
17620 |
0.14 |
| chr7_72929480_72929631 | 0.19 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
7047 |
0.15 |
| chr5_156644620_156644840 | 0.19 |
CTB-4E7.1 |
|
6660 |
0.14 |
| chr3_170995564_170995715 | 0.19 |
TNIK |
TRAF2 and NCK interacting kinase |
52139 |
0.17 |
| chr6_26174520_26174671 | 0.19 |
HIST1H2BE |
histone cluster 1, H2be |
9363 |
0.06 |
| chr13_43562200_43562351 | 0.19 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
3092 |
0.34 |
| chr3_56832571_56832900 | 0.19 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
3242 |
0.36 |
| chr1_228928769_228929191 | 0.19 |
RHOU |
ras homolog family member U |
58156 |
0.09 |
| chr6_42008817_42009180 | 0.19 |
CCND3 |
cyclin D3 |
7426 |
0.16 |
| chr14_91668757_91668960 | 0.19 |
C14orf159 |
chromosome 14 open reading frame 159 |
6121 |
0.19 |
| chr12_104912220_104912483 | 0.19 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
61572 |
0.13 |
| chr10_120973836_120973987 | 0.19 |
ENSG00000242853 |
. |
1542 |
0.31 |
| chr22_23190435_23190586 | 0.19 |
IGLV3-6 |
immunoglobulin lambda variable 3-6 (pseudogene) |
2040 |
0.09 |
| chr14_69090040_69090191 | 0.18 |
CTD-2325P2.4 |
|
5047 |
0.29 |
| chr1_160535898_160536088 | 0.18 |
CD84 |
CD84 molecule |
13270 |
0.13 |
| chr15_75159560_75159711 | 0.18 |
SCAMP2 |
secretory carrier membrane protein 2 |
5746 |
0.12 |
| chr11_12179023_12179437 | 0.18 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
3760 |
0.32 |
| chr4_76859668_76859847 | 0.18 |
NAAA |
N-acylethanolamine acid amidase |
1656 |
0.28 |
| chr12_15715155_15715553 | 0.18 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
16068 |
0.26 |
| chr11_67212387_67212841 | 0.18 |
CORO1B |
coronin, actin binding protein, 1B |
1351 |
0.18 |
| chr16_5022975_5023126 | 0.18 |
PPL |
periplakin |
12308 |
0.14 |
| chr11_47287617_47287768 | 0.18 |
MADD |
MAP-kinase activating death domain |
3020 |
0.14 |
| chr5_96341021_96341172 | 0.18 |
ENSG00000200884 |
. |
42635 |
0.13 |
| chr17_62965440_62965774 | 0.18 |
AMZ2P1 |
archaelysin family metallopeptidase 2 pseudogene 1 |
4028 |
0.17 |
| chr17_55519809_55520223 | 0.18 |
ENSG00000263902 |
. |
30454 |
0.2 |
| chr1_28525263_28525593 | 0.18 |
AL353354.2 |
|
1642 |
0.2 |
| chr14_89632758_89632909 | 0.18 |
FOXN3 |
forkhead box N3 |
14255 |
0.28 |
| chr1_39658074_39658238 | 0.18 |
MACF1 |
microtubule-actin crosslinking factor 1 |
11762 |
0.17 |
| chr13_25171578_25171729 | 0.18 |
ENSG00000211508 |
. |
11564 |
0.18 |
| chr1_39572189_39572340 | 0.18 |
MACF1 |
microtubule-actin crosslinking factor 1 |
1181 |
0.46 |
| chr12_53765461_53765679 | 0.18 |
SP1 |
Sp1 transcription factor |
8390 |
0.12 |
| chr1_100917512_100917831 | 0.18 |
RP5-837M10.4 |
|
33882 |
0.17 |
| chr17_40859652_40859819 | 0.18 |
CTD-3193K9.3 |
|
8251 |
0.09 |
| chr7_44666023_44666225 | 0.18 |
OGDH |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
2063 |
0.32 |
| chr16_68326071_68326222 | 0.18 |
ENSG00000252026 |
. |
7925 |
0.08 |
| chr14_52753608_52753759 | 0.18 |
PTGDR |
prostaglandin D2 receptor (DP) |
19153 |
0.22 |
| chr8_66913743_66914032 | 0.18 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
19908 |
0.26 |
| chr7_50302307_50302807 | 0.18 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
41767 |
0.19 |
| chr4_55543624_55543775 | 0.18 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
19614 |
0.29 |
| chr16_50874827_50875162 | 0.18 |
ENSG00000199771 |
. |
28492 |
0.15 |
| chr17_37914078_37914229 | 0.18 |
GRB7 |
growth factor receptor-bound protein 7 |
15648 |
0.11 |
| chr5_70884126_70884318 | 0.18 |
MCCC2 |
methylcrotonoyl-CoA carboxylase 2 (beta) |
1027 |
0.62 |
| chr1_26704569_26704720 | 0.18 |
ZNF683 |
zinc finger protein 683 |
3631 |
0.15 |
| chr10_98624657_98624808 | 0.18 |
LCOR |
ligand dependent nuclear receptor corepressor |
31995 |
0.15 |
| chr14_50507439_50508114 | 0.17 |
ENSG00000201358 |
. |
27510 |
0.13 |
| chr22_40349969_40350170 | 0.17 |
RP3-370M22.8 |
|
6332 |
0.18 |
| chr10_27145692_27145938 | 0.17 |
ABI1 |
abl-interactor 1 |
3977 |
0.23 |
| chr3_114051977_114052128 | 0.17 |
ENSG00000207770 |
. |
16636 |
0.16 |
| chr14_99688948_99689139 | 0.17 |
AL109767.1 |
|
40242 |
0.16 |
| chr14_35884998_35885149 | 0.17 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
11121 |
0.26 |
| chr14_98677293_98677464 | 0.17 |
ENSG00000222066 |
. |
120709 |
0.06 |
| chr2_231517982_231518133 | 0.17 |
CAB39 |
calcium binding protein 39 |
59503 |
0.11 |
| chr5_87436602_87437127 | 0.17 |
TMEM161B |
transmembrane protein 161B |
79584 |
0.11 |
| chr11_35180171_35180536 | 0.17 |
CD44 |
CD44 molecule (Indian blood group) |
17765 |
0.15 |
| chr3_133210635_133210930 | 0.17 |
BFSP2-AS1 |
BFSP2 antisense RNA 1 |
36162 |
0.16 |
| chr2_193045009_193045160 | 0.17 |
TMEFF2 |
transmembrane protein with EGF-like and two follistatin-like domains 2 |
14166 |
0.3 |
| chr17_26206098_26206388 | 0.17 |
LYRM9 |
LYR motif containing 9 |
4080 |
0.19 |
| chr18_20629843_20630057 | 0.17 |
ENSG00000223023 |
. |
25391 |
0.15 |
| chr1_198509908_198510168 | 0.17 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
37 |
0.99 |
| chr5_43397749_43397909 | 0.17 |
CCL28 |
chemokine (C-C motif) ligand 28 |
463 |
0.84 |
| chr17_16322494_16322752 | 0.17 |
TRPV2 |
transient receptor potential cation channel, subfamily V, member 2 |
28 |
0.96 |
| chr8_59974482_59974633 | 0.17 |
RP11-25K19.1 |
|
57042 |
0.12 |
| chr2_218811497_218811648 | 0.17 |
TNS1 |
tensin 1 |
2721 |
0.31 |
| chr20_50019481_50019826 | 0.17 |
ENSG00000263645 |
. |
25795 |
0.22 |
| chr12_31231062_31231213 | 0.17 |
DDX11 |
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
317 |
0.82 |
| chr7_149565914_149566574 | 0.17 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
3813 |
0.19 |
| chr1_246984990_246985280 | 0.17 |
ENSG00000252011 |
. |
11696 |
0.16 |
| chr6_37672753_37673067 | 0.17 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
5828 |
0.28 |
| chr3_4881435_4881664 | 0.17 |
ENSG00000239126 |
. |
38837 |
0.16 |
| chrX_117644722_117644873 | 0.17 |
DOCK11 |
dedicator of cytokinesis 11 |
14925 |
0.24 |
| chr11_32444065_32444216 | 0.17 |
WT1 |
Wilms tumor 1 |
4997 |
0.26 |
| chr2_74518143_74518523 | 0.17 |
SLC4A5 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
501 |
0.75 |
| chr3_188041188_188041339 | 0.17 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
83611 |
0.1 |
| chr12_12116177_12116328 | 0.17 |
ETV6 |
ets variant 6 |
77381 |
0.1 |
| chr7_156744058_156744240 | 0.17 |
NOM1 |
nucleolar protein with MIF4G domain 1 |
1732 |
0.36 |
| chr13_25200724_25201013 | 0.17 |
ENSG00000211508 |
. |
17651 |
0.18 |
| chr22_40915359_40915510 | 0.17 |
RP4-591N18.2 |
|
2370 |
0.26 |
| chr11_11158749_11159001 | 0.17 |
RP11-567I13.1 |
|
215098 |
0.02 |
| chr1_160605831_160606299 | 0.17 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
10746 |
0.16 |
| chr1_203285551_203285702 | 0.17 |
ENSG00000202300 |
. |
2601 |
0.25 |
| chr3_56831512_56831764 | 0.17 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
4339 |
0.33 |
| chr12_47588304_47588625 | 0.17 |
ENSG00000263838 |
. |
6869 |
0.23 |
| chr11_14436549_14436842 | 0.17 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
50643 |
0.14 |
| chr10_96163536_96163687 | 0.17 |
TBC1D12 |
TBC1 domain family, member 12 |
1350 |
0.49 |
| chr20_55204811_55205013 | 0.17 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
378 |
0.84 |
| chr2_198066929_198067080 | 0.17 |
ANKRD44 |
ankyrin repeat domain 44 |
4242 |
0.23 |
| chr2_46089761_46089912 | 0.17 |
PRKCE |
protein kinase C, epsilon |
138205 |
0.05 |
| chr17_66251665_66252057 | 0.17 |
ARSG |
arylsulfatase G |
3462 |
0.19 |
| chr5_35618942_35619093 | 0.16 |
SPEF2 |
sperm flagellar 2 |
917 |
0.65 |
| chr20_23620821_23620972 | 0.16 |
CST3 |
cystatin C |
1786 |
0.34 |
| chr16_9010574_9010725 | 0.16 |
USP7 |
ubiquitin specific peptidase 7 (herpes virus-associated) |
19064 |
0.15 |
| chr15_64916469_64916762 | 0.16 |
ENSG00000207223 |
. |
28472 |
0.13 |
| chrX_99880234_99880385 | 0.16 |
TSPAN6 |
tetraspanin 6 |
11494 |
0.18 |
| chr8_105634588_105634739 | 0.16 |
RP11-127H5.1 |
Uncharacterized protein |
31702 |
0.18 |
| chr4_83135377_83135528 | 0.16 |
ENSG00000202485 |
. |
39752 |
0.19 |
| chr15_101135936_101136223 | 0.16 |
LINS |
lines homolog (Drosophila) |
1062 |
0.44 |
| chr8_97175837_97176070 | 0.16 |
GDF6 |
growth differentiation factor 6 |
2933 |
0.31 |
| chr16_56296813_56297090 | 0.16 |
ENSG00000265281 |
. |
17519 |
0.16 |
| chr17_46911451_46911733 | 0.16 |
CALCOCO2 |
calcium binding and coiled-coil domain 2 |
3002 |
0.17 |
| chr19_35867894_35868045 | 0.16 |
GPR42 |
G protein-coupled receptor 42 (gene/pseudogene) |
5777 |
0.11 |
| chr20_36322114_36322265 | 0.16 |
CTNNBL1 |
catenin, beta like 1 |
219 |
0.96 |
| chr8_8195680_8195853 | 0.16 |
SGK223 |
Tyrosine-protein kinase SgK223 |
43491 |
0.16 |
| chr6_1895461_1895612 | 0.16 |
GMDS |
GDP-mannose 4,6-dehydratase |
280689 |
0.01 |
| chr4_15796755_15796906 | 0.16 |
CD38 |
CD38 molecule |
16929 |
0.23 |
| chr5_55980507_55980731 | 0.16 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
78560 |
0.09 |
| chr9_101789506_101789792 | 0.16 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
76671 |
0.09 |
| chr1_22414609_22414772 | 0.16 |
CDC42-IT1 |
CDC42 intronic transcript 1 (non-protein coding) |
29000 |
0.12 |
| chr10_6087510_6087742 | 0.16 |
IL2RA |
interleukin 2 receptor, alpha |
16627 |
0.13 |
| chr10_116261811_116262124 | 0.16 |
ABLIM1 |
actin binding LIM protein 1 |
14215 |
0.25 |
| chr1_157102658_157102809 | 0.16 |
ETV3 |
ets variant 3 |
5426 |
0.24 |
| chr3_68106098_68106274 | 0.16 |
FAM19A1 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
50813 |
0.19 |
| chr1_197745376_197745655 | 0.16 |
DENND1B |
DENN/MADD domain containing 1B |
752 |
0.71 |
| chr14_76063633_76063784 | 0.16 |
ENSG00000201096 |
. |
6808 |
0.16 |
| chr8_59961070_59961221 | 0.16 |
RP11-328K2.1 |
|
55648 |
0.13 |
| chr9_100689273_100689424 | 0.16 |
C9orf156 |
chromosome 9 open reading frame 156 |
4496 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.2 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.3 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.2 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.1 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0002669 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0072224 | metanephric glomerulus development(GO:0072224) metanephric glomerulus vasculature development(GO:0072239) |
| 0.0 | 0.1 | GO:0072133 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.1 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0043379 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.1 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.0 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) membrane raft localization(GO:0051665) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.0 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:1903170 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.0 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 | 0.1 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:1903313 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:0070602 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0008105 | asymmetric protein localization(GO:0008105) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0051023 | immunoglobulin secretion(GO:0048305) regulation of immunoglobulin secretion(GO:0051023) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.0 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0090079 | translation activator activity(GO:0008494) translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0035004 | phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.0 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.0 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |