Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
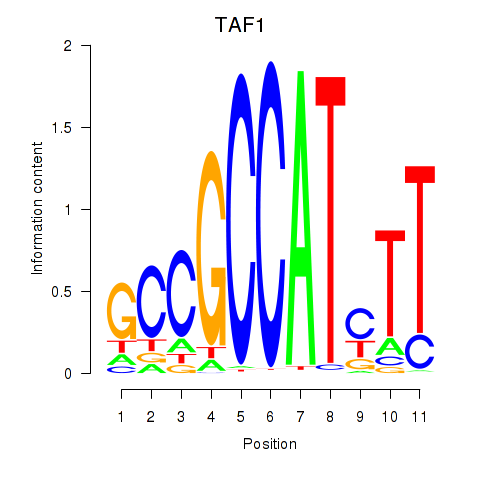
Results for TAF1
Z-value: 1.05
Transcription factors associated with TAF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TAF1
|
ENSG00000147133.11 | TATA-box binding protein associated factor 1 |
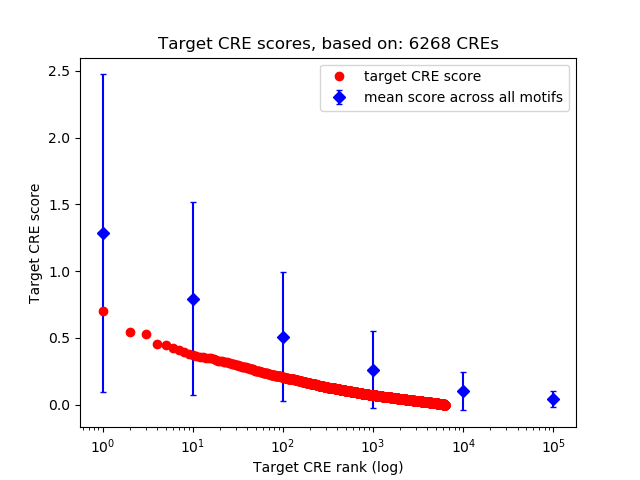
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_70587257_70587502 | TAF1 | 1224 | 0.488390 | 0.67 | 5.0e-02 | Click! |
| chrX_70587018_70587169 | TAF1 | 938 | 0.590509 | 0.64 | 6.3e-02 | Click! |
| chrX_70585571_70585723 | TAF1 | 467 | 0.825361 | 0.43 | 2.4e-01 | Click! |
| chrX_70585086_70585414 | TAF1 | 864 | 0.621556 | 0.39 | 3.1e-01 | Click! |
| chrX_70602910_70603061 | TAF1 | 10973 | 0.196501 | 0.35 | 3.6e-01 | Click! |
Activity of the TAF1 motif across conditions
Conditions sorted by the z-value of the TAF1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
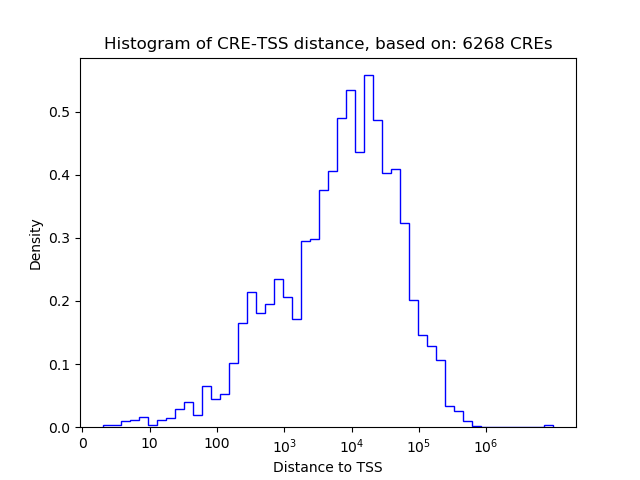
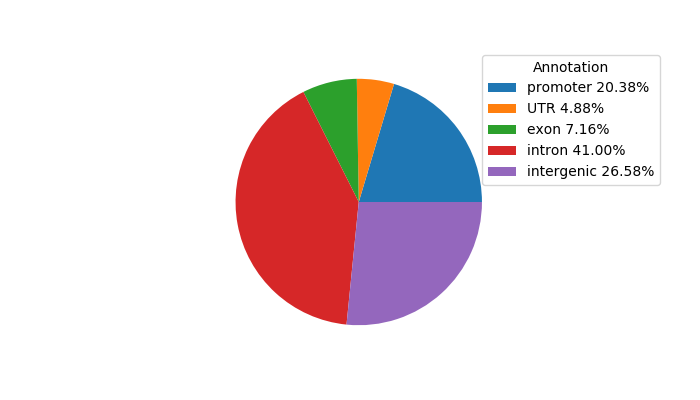
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_72736547_72736875 | 0.70 |
RAB37 |
RAB37, member RAS oncogene family |
3340 |
0.13 |
| chr1_40529943_40530230 | 0.55 |
PPT1 |
palmitoyl-protein thioesterase 1 |
17907 |
0.17 |
| chr1_151131154_151131481 | 0.53 |
SCNM1 |
sodium channel modifier 1 |
2177 |
0.13 |
| chr20_44626809_44627026 | 0.46 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
10630 |
0.12 |
| chr19_2628984_2629135 | 0.45 |
CTC-265F19.3 |
|
2893 |
0.19 |
| chr1_81674628_81674839 | 0.43 |
ENSG00000223026 |
. |
42741 |
0.2 |
| chr3_186502213_186502364 | 0.41 |
EIF4A2 |
eukaryotic translation initiation factor 4A2 |
296 |
0.31 |
| chr1_31223610_31223819 | 0.39 |
LAPTM5 |
lysosomal protein transmembrane 5 |
6953 |
0.17 |
| chr2_106416064_106416453 | 0.38 |
NCK2 |
NCK adaptor protein 2 |
16757 |
0.26 |
| chr1_111749620_111749829 | 0.37 |
DENND2D |
DENN/MADD domain containing 2D |
2693 |
0.18 |
| chr16_50700509_50700773 | 0.36 |
RP11-401P9.5 |
|
519 |
0.71 |
| chr12_117499069_117499579 | 0.36 |
TESC |
tescalcin |
14688 |
0.25 |
| chr17_32569270_32569530 | 0.36 |
CCL2 |
chemokine (C-C motif) ligand 2 |
12904 |
0.13 |
| chr2_103033478_103033629 | 0.35 |
IL18RAP |
interleukin 18 receptor accessory protein |
1596 |
0.31 |
| chr14_68162102_68162376 | 0.35 |
RDH11 |
retinol dehydrogenase 11 (all-trans/9-cis/11-cis) |
208 |
0.88 |
| chr9_71611720_71611963 | 0.35 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
17198 |
0.21 |
| chr5_137514277_137514428 | 0.34 |
BRD8 |
bromodomain containing 8 |
6 |
0.52 |
| chr2_38849799_38849986 | 0.34 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
19714 |
0.16 |
| chr12_104872773_104873085 | 0.33 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
22150 |
0.24 |
| chr8_61912101_61912451 | 0.33 |
CLVS1 |
clavesin 1 |
57441 |
0.16 |
| chr17_73649337_73649560 | 0.33 |
SMIM6 |
small integral membrane protein 6 |
6931 |
0.1 |
| chr2_27110608_27110889 | 0.32 |
DPYSL5 |
dihydropyrimidinase-like 5 |
39372 |
0.12 |
| chr1_208046544_208046843 | 0.32 |
CD34 |
CD34 molecule |
17144 |
0.25 |
| chr7_73643794_73644188 | 0.32 |
LAT2 |
linker for activation of T cells family, member 2 |
19638 |
0.13 |
| chr15_63788084_63788514 | 0.31 |
USP3 |
ubiquitin specific peptidase 3 |
8494 |
0.24 |
| chrX_71317772_71317960 | 0.31 |
RGAG4 |
retrotransposon gag domain containing 4 |
33812 |
0.14 |
| chr5_148928879_148929268 | 0.31 |
CSNK1A1 |
casein kinase 1, alpha 1 |
789 |
0.57 |
| chr16_30915705_30915976 | 0.31 |
CTF1 |
cardiotrophin 1 |
7881 |
0.08 |
| chr17_42846253_42846404 | 0.30 |
ADAM11 |
ADAM metallopeptidase domain 11 |
9635 |
0.14 |
| chr17_76144856_76145129 | 0.30 |
C17orf99 |
chromosome 17 open reading frame 99 |
2057 |
0.19 |
| chr12_93772804_93773052 | 0.30 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
498 |
0.69 |
| chr9_90229654_90229805 | 0.29 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
61360 |
0.12 |
| chr1_66862492_66862643 | 0.29 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
42502 |
0.2 |
| chr11_67368590_67368774 | 0.29 |
RP11-655M14.12 |
|
4310 |
0.1 |
| chr6_6799100_6799314 | 0.28 |
ENSG00000240936 |
. |
139923 |
0.05 |
| chrX_48796338_48796526 | 0.28 |
OTUD5 |
OTU domain containing 5 |
18019 |
0.08 |
| chr1_848466_848694 | 0.28 |
SAMD11 |
sterile alpha motif domain containing 11 |
11680 |
0.11 |
| chr21_34757403_34757693 | 0.28 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
17654 |
0.17 |
| chr14_51251627_51251873 | 0.28 |
NIN |
ninein (GSK3B interacting protein) |
18237 |
0.15 |
| chr1_88748973_88749156 | 0.28 |
ENSG00000239504 |
. |
194733 |
0.03 |
| chr2_43438738_43438979 | 0.27 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
14890 |
0.22 |
| chr3_172235975_172236263 | 0.27 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
5146 |
0.28 |
| chr3_31549610_31550078 | 0.27 |
STT3B |
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
24438 |
0.26 |
| chrX_48334189_48334340 | 0.27 |
FTSJ1 |
FtsJ RNA methyltransferase homolog 1 (E. coli) |
285 |
0.84 |
| chr9_134851506_134851657 | 0.27 |
MED27 |
mediator complex subunit 27 |
103672 |
0.07 |
| chr4_122744076_122744227 | 0.27 |
CCNA2 |
cyclin A2 |
936 |
0.53 |
| chr1_175157259_175157701 | 0.27 |
KIAA0040 |
KIAA0040 |
4410 |
0.3 |
| chr7_82072704_82072883 | 0.26 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
238 |
0.96 |
| chr5_150284385_150284536 | 0.25 |
ZNF300 |
zinc finger protein 300 |
80 |
0.97 |
| chr2_134991809_134992312 | 0.25 |
ENSG00000222921 |
. |
3294 |
0.31 |
| chr9_72069887_72070175 | 0.25 |
RP11-470P21.2 |
|
17351 |
0.25 |
| chr11_46341833_46341984 | 0.25 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
9200 |
0.16 |
| chr13_42622723_42622874 | 0.25 |
DGKH |
diacylglycerol kinase, eta |
91 |
0.97 |
| chr15_91459819_91459977 | 0.25 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
3780 |
0.1 |
| chr7_6179339_6179551 | 0.25 |
USP42 |
ubiquitin specific peptidase 42 |
23334 |
0.12 |
| chr12_6572982_6573330 | 0.25 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
6655 |
0.09 |
| chr10_28656729_28657019 | 0.24 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
33459 |
0.16 |
| chr1_151247807_151247958 | 0.24 |
ZNF687 |
zinc finger protein 687 |
6212 |
0.1 |
| chr4_102203836_102204080 | 0.24 |
ENSG00000221265 |
. |
47613 |
0.15 |
| chr12_101673837_101673988 | 0.24 |
UTP20 |
UTP20, small subunit (SSU) processome component, homolog (yeast) |
25 |
0.98 |
| chr14_35183530_35183681 | 0.24 |
CFL2 |
cofilin 2 (muscle) |
156 |
0.96 |
| chr11_87063041_87063296 | 0.24 |
ENSG00000223015 |
. |
254507 |
0.02 |
| chr1_77881942_77882095 | 0.24 |
ENSG00000251767 |
. |
3992 |
0.26 |
| chr14_92986394_92986545 | 0.24 |
RIN3 |
Ras and Rab interactor 3 |
6321 |
0.3 |
| chr10_81009151_81009302 | 0.24 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
56749 |
0.14 |
| chr15_59397716_59397952 | 0.24 |
CCNB2 |
cyclin B2 |
478 |
0.74 |
| chrX_15932844_15932995 | 0.23 |
ENSG00000200566 |
. |
1507 |
0.49 |
| chrX_47225759_47225910 | 0.23 |
ZNF157 |
zinc finger protein 157 |
4148 |
0.18 |
| chr12_133240338_133240655 | 0.23 |
POLE |
polymerase (DNA directed), epsilon, catalytic subunit |
2997 |
0.19 |
| chr6_111177453_111177656 | 0.23 |
ENSG00000199360 |
. |
66 |
0.97 |
| chr17_61497054_61497266 | 0.23 |
AC015923.1 |
|
5487 |
0.14 |
| chr19_23432669_23432887 | 0.23 |
ZNF724P |
zinc finger protein 724, pseudogene |
384 |
0.9 |
| chr16_75662277_75662523 | 0.23 |
ADAT1 |
adenosine deaminase, tRNA-specific 1 |
5202 |
0.15 |
| chr16_81510608_81510759 | 0.23 |
CMIP |
c-Maf inducing protein |
18271 |
0.25 |
| chr11_67178315_67178466 | 0.23 |
CARNS1 |
carnosine synthase 1 |
4759 |
0.07 |
| chr17_74143785_74143956 | 0.22 |
RNF157-AS1 |
RNF157 antisense RNA 1 |
5923 |
0.12 |
| chr12_94520855_94521052 | 0.22 |
PLXNC1 |
plexin C1 |
21546 |
0.21 |
| chr4_53525238_53525389 | 0.22 |
USP46 |
ubiquitin specific peptidase 46 |
144 |
0.96 |
| chr14_94624184_94624480 | 0.22 |
PPP4R4 |
protein phosphatase 4, regulatory subunit 4 |
11867 |
0.17 |
| chr3_30685597_30685782 | 0.22 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
37596 |
0.2 |
| chr2_61763268_61763781 | 0.22 |
XPO1 |
exportin 1 (CRM1 homolog, yeast) |
199 |
0.94 |
| chr19_23941207_23941438 | 0.22 |
ZNF681 |
zinc finger protein 681 |
317 |
0.89 |
| chr7_72791080_72791282 | 0.22 |
ENSG00000202035 |
. |
27038 |
0.12 |
| chr20_58632008_58632274 | 0.22 |
C20orf197 |
chromosome 20 open reading frame 197 |
1161 |
0.58 |
| chr2_69004064_69004400 | 0.22 |
ARHGAP25 |
Rho GTPase activating protein 25 |
2160 |
0.38 |
| chr1_169014315_169014663 | 0.22 |
ENSG00000252987 |
. |
22013 |
0.19 |
| chr20_61435133_61435481 | 0.22 |
OGFR |
opioid growth factor receptor |
880 |
0.4 |
| chr4_775014_775218 | 0.22 |
RP11-440L14.1 |
|
502 |
0.7 |
| chr20_20687843_20687994 | 0.21 |
RALGAPA2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
5213 |
0.29 |
| chr10_6342998_6343158 | 0.21 |
DKFZP667F0711 |
|
49200 |
0.13 |
| chr8_94753070_94753221 | 0.21 |
RBM12B |
RNA binding motif protein 12B |
39 |
0.75 |
| chr4_110604141_110604292 | 0.21 |
AC004067.5 |
|
8944 |
0.19 |
| chrX_131119863_131120181 | 0.21 |
ENSG00000265686 |
. |
10851 |
0.22 |
| chr1_65524057_65524208 | 0.21 |
ENSG00000199135 |
. |
59 |
0.92 |
| chr10_129853482_129853633 | 0.21 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
7723 |
0.29 |
| chr11_34886337_34886667 | 0.21 |
PDHX |
pyruvate dehydrogenase complex, component X |
50874 |
0.14 |
| chr16_68109101_68109477 | 0.21 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
9958 |
0.1 |
| chr18_42645785_42646163 | 0.21 |
ENSG00000265957 |
. |
95843 |
0.08 |
| chr6_144502256_144502416 | 0.21 |
STX11 |
syntaxin 11 |
30673 |
0.21 |
| chr5_162833953_162834104 | 0.21 |
RP11-541P9.3 |
|
30271 |
0.13 |
| chr5_173313190_173313341 | 0.21 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
2018 |
0.44 |
| chr19_4670232_4670383 | 0.20 |
C19orf10 |
chromosome 19 open reading frame 10 |
67 |
0.95 |
| chr2_231362586_231362737 | 0.20 |
SP100 |
SP100 nuclear antigen |
6316 |
0.21 |
| chr9_116351624_116351848 | 0.20 |
RP11-168K11.2 |
|
563 |
0.78 |
| chr10_45571216_45571367 | 0.20 |
RSU1P2 |
Ras suppressor protein 1 pseudogene 2 |
40258 |
0.13 |
| chr19_21949976_21950127 | 0.20 |
ZNF100 |
zinc finger protein 100 |
300 |
0.91 |
| chr11_46343510_46343716 | 0.20 |
DGKZ |
diacylglycerol kinase, zeta |
10842 |
0.15 |
| chr16_79126121_79126272 | 0.20 |
RP11-556H2.3 |
|
2249 |
0.3 |
| chr7_73102787_73102938 | 0.20 |
WBSCR22 |
Williams Beuren syndrome chromosome region 22 |
2384 |
0.17 |
| chr4_139335299_139335467 | 0.20 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
171880 |
0.04 |
| chr2_8630914_8631065 | 0.20 |
AC011747.7 |
|
184907 |
0.03 |
| chr20_42698514_42698843 | 0.20 |
TOX2 |
TOX high mobility group box family member 2 |
15687 |
0.22 |
| chr19_24216546_24216697 | 0.20 |
ZNF254 |
zinc finger protein 254 |
345 |
0.9 |
| chr17_37049047_37049226 | 0.20 |
RP1-56K13.2 |
|
11717 |
0.1 |
| chr2_128106575_128106780 | 0.20 |
MAP3K2 |
mitogen-activated protein kinase kinase kinase 2 |
5872 |
0.19 |
| chrX_117637530_117637707 | 0.19 |
DOCK11 |
dedicator of cytokinesis 11 |
7746 |
0.26 |
| chr2_43018765_43019093 | 0.19 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
783 |
0.7 |
| chr17_45770162_45770366 | 0.19 |
TBKBP1 |
TBK1 binding protein 1 |
1183 |
0.38 |
| chr12_111471925_111472076 | 0.19 |
CUX2 |
cut-like homeobox 2 |
172 |
0.96 |
| chr7_38401901_38402261 | 0.19 |
AMPH |
amphiphysin |
100632 |
0.08 |
| chr2_152494355_152494506 | 0.19 |
NEB |
nebulin |
54297 |
0.17 |
| chr10_12286201_12286352 | 0.19 |
RP11-186N15.3 |
|
1881 |
0.27 |
| chr12_105077721_105077931 | 0.19 |
ENSG00000264295 |
. |
92415 |
0.08 |
| chr7_2685977_2686128 | 0.19 |
TTYH3 |
tweety family member 3 |
886 |
0.6 |
| chr21_39844669_39844820 | 0.19 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
25601 |
0.26 |
| chr15_70751898_70752049 | 0.19 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
242647 |
0.02 |
| chr3_71757856_71758018 | 0.19 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
16589 |
0.21 |
| chr21_39879302_39879453 | 0.19 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
8970 |
0.31 |
| chr5_32211061_32211425 | 0.19 |
ENSG00000199731 |
. |
23738 |
0.17 |
| chrX_73071562_73071713 | 0.19 |
RP13-216E22.5 |
|
117683 |
0.06 |
| chr4_75960540_75960691 | 0.19 |
RP11-44F21.2 |
|
581 |
0.83 |
| chr16_89041472_89041726 | 0.19 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
1802 |
0.3 |
| chr3_111262708_111262863 | 0.19 |
CD96 |
CD96 molecule |
1788 |
0.45 |
| chr9_6942449_6942685 | 0.19 |
KDM4C |
lysine (K)-specific demethylase 4C |
17066 |
0.25 |
| chr2_73259129_73259280 | 0.19 |
SFXN5 |
sideroflexin 5 |
39598 |
0.15 |
| chr1_90222717_90222959 | 0.19 |
ENSG00000239176 |
. |
11039 |
0.21 |
| chr19_23258312_23258470 | 0.19 |
ZNF730 |
zinc finger protein 730 |
379 |
0.91 |
| chr6_2793669_2793866 | 0.19 |
WRNIP1 |
Werner helicase interacting protein 1 |
24760 |
0.16 |
| chr1_111749000_111749179 | 0.19 |
DENND2D |
DENN/MADD domain containing 2D |
2058 |
0.22 |
| chr12_32006284_32006435 | 0.18 |
ENSG00000252204 |
. |
42337 |
0.12 |
| chr11_118748210_118748363 | 0.18 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
6189 |
0.11 |
| chr6_15086494_15086645 | 0.18 |
ENSG00000242989 |
. |
26630 |
0.23 |
| chr1_207994075_207994226 | 0.18 |
ENSG00000203709 |
. |
18282 |
0.22 |
| chr19_34307726_34307972 | 0.18 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
10020 |
0.28 |
| chr14_93034614_93034765 | 0.18 |
RIN3 |
Ras and Rab interactor 3 |
54541 |
0.16 |
| chr2_120142991_120143276 | 0.18 |
DBI |
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
17880 |
0.17 |
| chr9_137290760_137290930 | 0.18 |
RXRA |
retinoid X receptor, alpha |
7583 |
0.24 |
| chr11_34469458_34469624 | 0.18 |
CAT |
catalase |
9069 |
0.22 |
| chr7_64022966_64023226 | 0.18 |
ZNF680 |
zinc finger protein 680 |
323 |
0.94 |
| chr15_86100041_86100575 | 0.18 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
1631 |
0.38 |
| chr19_22235520_22235840 | 0.18 |
ZNF257 |
zinc finger protein 257 |
342 |
0.92 |
| chrX_12919691_12919842 | 0.18 |
TLR8 |
toll-like receptor 8 |
4973 |
0.21 |
| chr14_98173448_98173821 | 0.18 |
RP11-204N11.1 |
Uncharacterized protein |
55534 |
0.17 |
| chr17_76706821_76706987 | 0.18 |
CYTH1 |
cytohesin 1 |
6203 |
0.21 |
| chr19_52067696_52067847 | 0.18 |
ZNF175 |
zinc finger protein 175 |
6780 |
0.12 |
| chr3_196351016_196351281 | 0.18 |
LINC01063 |
long intergenic non-protein coding RNA 1063 |
8310 |
0.13 |
| chr15_52971703_52971888 | 0.18 |
FAM214A |
family with sequence similarity 214, member A |
226 |
0.95 |
| chr10_22611140_22611362 | 0.18 |
BMI1 |
BMI1 polycomb ring finger oncogene |
216 |
0.91 |
| chr2_233957419_233957649 | 0.18 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
32345 |
0.16 |
| chr18_74196362_74196513 | 0.18 |
ZNF516 |
zinc finger protein 516 |
6298 |
0.18 |
| chr11_95988623_95988872 | 0.17 |
ENSG00000266192 |
. |
85855 |
0.09 |
| chr12_31468309_31468579 | 0.17 |
FAM60A |
family with sequence similarity 60, member A |
8652 |
0.16 |
| chr2_54198442_54198698 | 0.17 |
ACYP2 |
acylphosphatase 2, muscle type |
325 |
0.69 |
| chr11_6766326_6766551 | 0.17 |
GVINP1 |
GTPase, very large interferon inducible pseudogene 1 |
23327 |
0.11 |
| chr1_161592599_161593034 | 0.17 |
FCGR3B |
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
8006 |
0.12 |
| chr10_72545418_72545569 | 0.17 |
TBATA |
thymus, brain and testes associated |
336 |
0.9 |
| chr14_92986657_92986808 | 0.17 |
RIN3 |
Ras and Rab interactor 3 |
6584 |
0.3 |
| chr8_27212033_27212272 | 0.17 |
PTK2B |
protein tyrosine kinase 2 beta |
26016 |
0.18 |
| chr9_111528485_111528636 | 0.17 |
ACTL7B |
actin-like 7B |
90679 |
0.08 |
| chr17_27263377_27264057 | 0.17 |
RP11-20B24.6 |
|
8097 |
0.09 |
| chr2_230787185_230787336 | 0.17 |
FBXO36 |
F-box protein 36 |
38 |
0.9 |
| chr12_54685962_54686189 | 0.17 |
NFE2 |
nuclear factor, erythroid 2 |
3467 |
0.1 |
| chr13_49432430_49432644 | 0.17 |
ENSG00000265585 |
. |
71811 |
0.11 |
| chr11_101982918_101983069 | 0.17 |
YAP1 |
Yes-associated protein 1 |
252 |
0.91 |
| chrX_13106186_13106576 | 0.17 |
FAM9C |
family with sequence similarity 9, member C |
43580 |
0.18 |
| chr15_40453377_40453695 | 0.17 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
268 |
0.9 |
| chr16_12103370_12103521 | 0.17 |
RP11-166B2.7 |
|
31738 |
0.1 |
| chr8_27248376_27248527 | 0.17 |
PTK2B |
protein tyrosine kinase 2 beta |
6518 |
0.25 |
| chr7_127669033_127669184 | 0.17 |
LRRC4 |
leucine rich repeat containing 4 |
1950 |
0.41 |
| chr3_45721073_45721330 | 0.17 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
9173 |
0.18 |
| chr11_85854528_85854819 | 0.17 |
ENSG00000200877 |
. |
9596 |
0.23 |
| chr5_131830049_131830573 | 0.17 |
IRF1 |
interferon regulatory factor 1 |
3821 |
0.17 |
| chr9_209184_209369 | 0.17 |
DOCK8 |
dedicator of cytokinesis 8 |
5589 |
0.19 |
| chr22_29771553_29771704 | 0.17 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
4980 |
0.16 |
| chr3_185970050_185970201 | 0.17 |
DGKG |
diacylglycerol kinase, gamma 90kDa |
28378 |
0.23 |
| chr5_171710215_171710366 | 0.16 |
UBTD2 |
ubiquitin domain containing 2 |
785 |
0.61 |
| chr15_70451459_70451610 | 0.16 |
ENSG00000200216 |
. |
34041 |
0.2 |
| chr8_61151340_61151491 | 0.16 |
CA8 |
carbonic anhydrase VIII |
42556 |
0.22 |
| chr6_159489783_159489978 | 0.16 |
TAGAP |
T-cell activation RhoGTPase activating protein |
23696 |
0.18 |
| chr7_64466611_64466762 | 0.16 |
ERV3-1 |
endogenous retrovirus group 3, member 1 |
345 |
0.85 |
| chr3_71253678_71253850 | 0.16 |
FOXP1 |
forkhead box P1 |
6232 |
0.32 |
| chr4_26077549_26077700 | 0.16 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
87453 |
0.1 |
| chr2_243034231_243034506 | 0.16 |
AC093642.5 |
|
2677 |
0.29 |
| chr3_192635217_192635637 | 0.16 |
MB21D2 |
Mab-21 domain containing 2 |
523 |
0.87 |
| chr22_47253139_47253290 | 0.16 |
ENSG00000221672 |
. |
9411 |
0.26 |
| chr19_14227765_14227948 | 0.16 |
PRKACA |
protein kinase, cAMP-dependent, catalytic, alpha |
688 |
0.47 |
| chr10_70480665_70480816 | 0.16 |
CCAR1 |
cell division cycle and apoptosis regulator 1 |
29 |
0.96 |
| chr11_117843552_117843785 | 0.16 |
IL10RA |
interleukin 10 receptor, alpha |
13395 |
0.17 |
| chr1_117283826_117284150 | 0.16 |
CD2 |
CD2 molecule |
13019 |
0.2 |
| chrX_65240625_65241043 | 0.16 |
ENSG00000207939 |
. |
2122 |
0.33 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.2 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.1 | 0.3 | GO:0002517 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.1 | 0.2 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.2 | GO:0035751 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.2 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.0 | GO:0045588 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.2 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.3 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.0 | 0.1 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.0 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.0 | GO:0042754 | negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.2 | GO:0075733 | multi-organism transport(GO:0044766) transport of virus(GO:0046794) intracellular transport of virus(GO:0075733) multi-organism localization(GO:1902579) multi-organism intracellular transport(GO:1902583) |
| 0.0 | 0.1 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.0 | GO:0002645 | regulation of tolerance induction(GO:0002643) positive regulation of tolerance induction(GO:0002645) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.0 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.0 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.1 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0090075 | relaxation of muscle(GO:0090075) |
| 0.0 | 0.1 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.0 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0001562 | response to protozoan(GO:0001562) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
| 0.0 | 0.3 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0043189 | H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.0 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.7 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |