Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
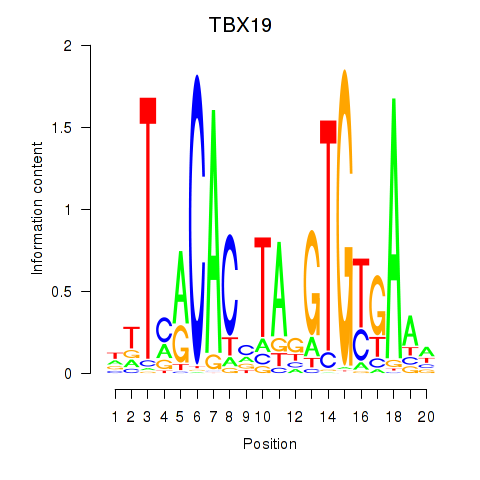
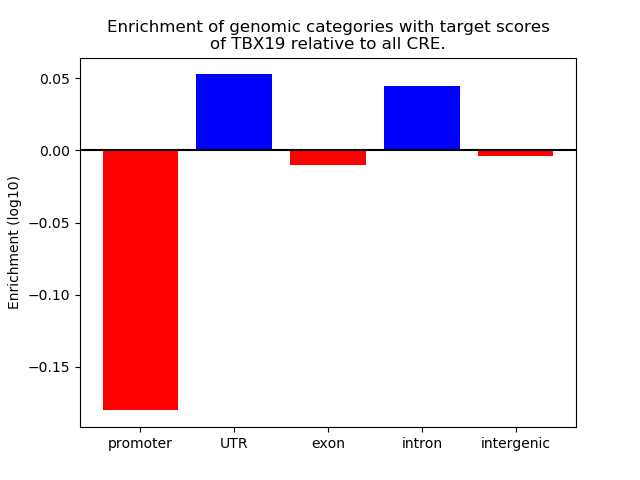
Results for TBX19
Z-value: 1.39
Transcription factors associated with TBX19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX19
|
ENSG00000143178.8 | T-box transcription factor 19 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_168290494_168290645 | TBX19 | 23707 | 0.189074 | -0.48 | 1.9e-01 | Click! |
| chr1_168289944_168290095 | TBX19 | 23157 | 0.190319 | -0.27 | 4.8e-01 | Click! |
| chr1_168271269_168271420 | TBX19 | 4482 | 0.246188 | -0.07 | 8.5e-01 | Click! |
| chr1_168254552_168254703 | TBX19 | 4349 | 0.233789 | -0.00 | 1.0e+00 | Click! |
Activity of the TBX19 motif across conditions
Conditions sorted by the z-value of the TBX19 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
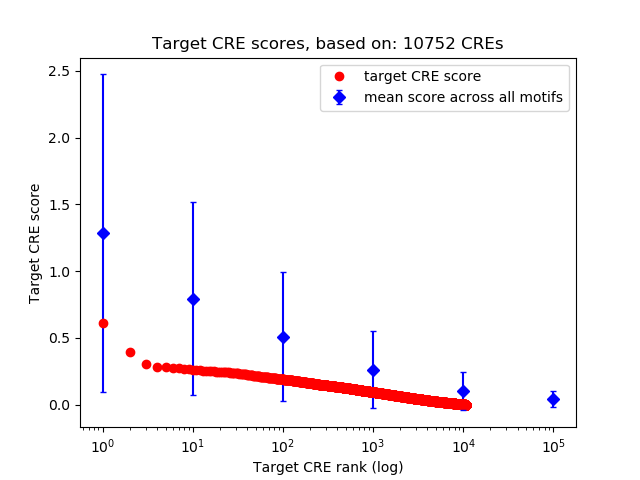
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_46530833_46530984 | 0.61 |
PGLYRP1 |
peptidoglycan recognition protein 1 |
4585 |
0.14 |
| chr20_13838142_13838293 | 0.40 |
NDUFAF5 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 |
72521 |
0.1 |
| chr5_141307102_141307397 | 0.30 |
KIAA0141 |
KIAA0141 |
3774 |
0.18 |
| chr3_121386721_121386872 | 0.29 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
7038 |
0.16 |
| chr14_65165014_65165274 | 0.28 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
5676 |
0.24 |
| chr15_70755158_70755354 | 0.28 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
239364 |
0.02 |
| chr21_43614804_43615118 | 0.28 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
4838 |
0.21 |
| chr10_82421596_82421747 | 0.27 |
SH2D4B |
SH2 domain containing 4B |
121096 |
0.06 |
| chr2_198150239_198150434 | 0.26 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
16907 |
0.16 |
| chr1_38958842_38958993 | 0.26 |
ENSG00000200796 |
. |
96586 |
0.08 |
| chr9_92272620_92272771 | 0.26 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
52742 |
0.17 |
| chr8_97662802_97662953 | 0.26 |
CPQ |
carboxypeptidase Q |
5274 |
0.31 |
| chr17_40994693_40994844 | 0.25 |
AOC2 |
amine oxidase, copper containing 2 (retina-specific) |
1849 |
0.16 |
| chr14_91167266_91167417 | 0.25 |
RP11-661G16.2 |
|
3664 |
0.23 |
| chr8_59068204_59068401 | 0.25 |
FAM110B |
family with sequence similarity 110, member B |
161189 |
0.04 |
| chr14_90032899_90033050 | 0.25 |
FOXN3-AS2 |
FOXN3 antisense RNA 2 |
9586 |
0.17 |
| chr2_118814167_118814318 | 0.25 |
INSIG2 |
insulin induced gene 2 |
31808 |
0.13 |
| chr1_212000871_212001169 | 0.25 |
RP11-552D8.1 |
|
2168 |
0.25 |
| chrX_134441517_134441668 | 0.25 |
ZNF75D |
zinc finger protein 75D |
11633 |
0.21 |
| chr15_86078641_86078893 | 0.25 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
8504 |
0.21 |
| chr20_31898220_31898371 | 0.25 |
BPIFB1 |
BPI fold containing family B, member 1 |
27354 |
0.14 |
| chr17_74246966_74247117 | 0.24 |
RNF157 |
ring finger protein 157 |
10587 |
0.13 |
| chr15_99488986_99489137 | 0.24 |
IGF1R |
insulin-like growth factor 1 receptor |
21232 |
0.19 |
| chr16_15883480_15883631 | 0.24 |
AF001548.6 |
|
48547 |
0.1 |
| chr18_74188097_74188669 | 0.24 |
ZNF516 |
zinc finger protein 516 |
14352 |
0.17 |
| chr2_144450054_144450205 | 0.24 |
RP11-434H14.1 |
|
9621 |
0.23 |
| chr4_184318912_184319063 | 0.24 |
ENSG00000263395 |
. |
13281 |
0.17 |
| chr20_58675900_58676156 | 0.24 |
C20orf197 |
chromosome 20 open reading frame 197 |
45048 |
0.18 |
| chr1_52105044_52105195 | 0.24 |
OSBPL9 |
oxysterol binding protein-like 9 |
22338 |
0.19 |
| chr18_43578628_43579079 | 0.23 |
ENSG00000222179 |
. |
8942 |
0.2 |
| chr1_32558443_32558736 | 0.23 |
ENSG00000266203 |
. |
6039 |
0.13 |
| chr21_15810558_15810709 | 0.23 |
HSPA13 |
heat shock protein 70kDa family, member 13 |
55125 |
0.15 |
| chr8_10028580_10028731 | 0.23 |
MSRA |
methionine sulfoxide reductase A |
75418 |
0.1 |
| chr3_196404146_196404677 | 0.23 |
ENSG00000201441 |
. |
31424 |
0.1 |
| chr3_131537149_131537300 | 0.23 |
CPNE4 |
copine IV |
107201 |
0.07 |
| chr2_202074857_202075031 | 0.23 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
23222 |
0.14 |
| chr1_180238065_180238216 | 0.23 |
RP5-1180C10.2 |
|
5676 |
0.25 |
| chr12_110435362_110435658 | 0.23 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
1316 |
0.37 |
| chr20_24270958_24271313 | 0.23 |
ENSG00000200231 |
. |
53244 |
0.17 |
| chrX_65232472_65232880 | 0.22 |
ENSG00000207939 |
. |
6036 |
0.22 |
| chr6_5828955_5829300 | 0.22 |
ENSG00000239472 |
. |
89950 |
0.09 |
| chr14_91592357_91592508 | 0.22 |
ENSG00000221102 |
. |
337 |
0.64 |
| chr15_42662964_42663115 | 0.22 |
CAPN3 |
calpain 3, (p94) |
11254 |
0.16 |
| chr20_35586845_35586996 | 0.22 |
SAMHD1 |
SAM domain and HD domain 1 |
6674 |
0.24 |
| chr18_74211925_74212216 | 0.22 |
RP11-17M16.1 |
uncharacterized protein LOC400658 |
4593 |
0.19 |
| chr22_22304135_22304332 | 0.22 |
PPM1F |
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
2970 |
0.16 |
| chr14_64214575_64214726 | 0.22 |
ENSG00000252749 |
. |
11488 |
0.18 |
| chr11_78109102_78109366 | 0.22 |
RP11-452H21.2 |
|
10127 |
0.18 |
| chr2_69063618_69063857 | 0.22 |
AC097495.2 |
|
1601 |
0.39 |
| chr3_71376865_71377143 | 0.22 |
FOXP1 |
forkhead box P1 |
23093 |
0.23 |
| chr6_131889933_131890084 | 0.22 |
ARG1 |
arginase 1 |
4276 |
0.25 |
| chr4_185772755_185773053 | 0.21 |
ENSG00000266698 |
. |
640 |
0.72 |
| chr1_172410212_172410374 | 0.21 |
PIGC |
phosphatidylinositol glycan anchor biosynthesis, class C |
2907 |
0.24 |
| chr9_95555847_95555998 | 0.21 |
ANKRD19P |
ankyrin repeat domain 19, pseudogene |
15971 |
0.16 |
| chr7_156842429_156842580 | 0.21 |
MNX1-AS1 |
MNX1 antisense RNA 1 (head to head) |
39005 |
0.12 |
| chr1_63931746_63931897 | 0.21 |
ITGB3BP |
integrin beta 3 binding protein (beta3-endonexin) |
57030 |
0.09 |
| chr2_42979198_42979349 | 0.21 |
OXER1 |
oxoeicosanoid (OXE) receptor 1 |
12128 |
0.21 |
| chr2_68652846_68653082 | 0.21 |
FBXO48 |
F-box protein 48 |
41426 |
0.12 |
| chr18_74807316_74807807 | 0.21 |
MBP |
myelin basic protein |
9656 |
0.29 |
| chr12_132366972_132367123 | 0.21 |
ULK1 |
unc-51 like autophagy activating kinase 1 |
12149 |
0.17 |
| chr6_116784453_116784778 | 0.21 |
RP1-93H18.6 |
|
240 |
0.83 |
| chr4_48239820_48240029 | 0.21 |
TEC |
tec protein tyrosine kinase |
31957 |
0.18 |
| chr17_4158247_4158398 | 0.21 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
8820 |
0.18 |
| chr2_207025046_207025348 | 0.21 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
866 |
0.28 |
| chr14_51292294_51292451 | 0.21 |
NIN |
ninein (GSK3B interacting protein) |
3598 |
0.17 |
| chr2_28843080_28843618 | 0.20 |
PLB1 |
phospholipase B1 |
10104 |
0.21 |
| chr3_154802157_154802308 | 0.20 |
MME |
membrane metallo-endopeptidase |
499 |
0.88 |
| chr4_79597785_79597936 | 0.20 |
ENSG00000238816 |
. |
36606 |
0.16 |
| chr1_93495068_93495341 | 0.20 |
ENSG00000251837 |
. |
2141 |
0.21 |
| chr1_234899568_234899784 | 0.20 |
ENSG00000201638 |
. |
74044 |
0.11 |
| chr2_198971191_198971372 | 0.20 |
PLCL1 |
phospholipase C-like 1 |
22745 |
0.28 |
| chr8_131021838_131021989 | 0.20 |
ENSG00000264653 |
. |
1214 |
0.43 |
| chr8_39806379_39806556 | 0.20 |
IDO2 |
indoleamine 2,3-dioxygenase 2 |
218 |
0.93 |
| chr12_57106930_57107081 | 0.20 |
NACA |
nascent polypeptide-associated complex alpha subunit |
6379 |
0.13 |
| chr2_239295864_239296015 | 0.20 |
ENSG00000202099 |
. |
25344 |
0.14 |
| chr2_128938668_128938819 | 0.20 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
1383 |
0.51 |
| chr1_152022964_152023546 | 0.20 |
S100A11 |
S100 calcium binding protein A11 |
13744 |
0.14 |
| chrX_119670283_119670616 | 0.20 |
ENSG00000272179 |
. |
7721 |
0.2 |
| chrX_12928349_12928500 | 0.20 |
TLR8-AS1 |
TLR8 antisense RNA 1 |
1972 |
0.31 |
| chr1_168416183_168416334 | 0.20 |
ENSG00000207974 |
. |
71496 |
0.11 |
| chr7_75505515_75505666 | 0.20 |
RHBDD2 |
rhomboid domain containing 2 |
2676 |
0.23 |
| chr3_151592281_151592432 | 0.20 |
SUCNR1 |
succinate receptor 1 |
925 |
0.64 |
| chr4_14913654_14913805 | 0.20 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
90569 |
0.1 |
| chr20_35470625_35470932 | 0.20 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
17498 |
0.15 |
| chr14_71434574_71434725 | 0.20 |
PCNX |
pecanex homolog (Drosophila) |
45099 |
0.19 |
| chr1_169572243_169572400 | 0.19 |
SELP |
selectin P (granule membrane protein 140kDa, antigen CD62) |
16136 |
0.19 |
| chr12_95032892_95033043 | 0.19 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
11371 |
0.26 |
| chr3_39291368_39291710 | 0.19 |
CX3CR1 |
chemokine (C-X3-C motif) receptor 1 |
29986 |
0.16 |
| chr20_4797995_4798146 | 0.19 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
2301 |
0.31 |
| chr8_124145367_124145518 | 0.19 |
TBC1D31 |
TBC1 domain family, member 31 |
32249 |
0.11 |
| chr22_17597863_17598182 | 0.19 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
4121 |
0.17 |
| chr1_60015308_60015459 | 0.19 |
FGGY |
FGGY carbohydrate kinase domain containing |
4207 |
0.36 |
| chr14_65200681_65201005 | 0.19 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
3890 |
0.26 |
| chr7_41525377_41525528 | 0.19 |
INHBA-AS1 |
INHBA antisense RNA 1 |
208062 |
0.03 |
| chr1_185390714_185391145 | 0.19 |
ENSG00000252407 |
. |
12447 |
0.26 |
| chr2_46116515_46116666 | 0.19 |
PRKCE |
protein kinase C, epsilon |
111451 |
0.07 |
| chr20_49073611_49073762 | 0.19 |
ENSG00000244376 |
. |
27666 |
0.17 |
| chr6_160363198_160363349 | 0.19 |
IGF2R |
insulin-like growth factor 2 receptor |
26858 |
0.17 |
| chr2_135953447_135953598 | 0.19 |
ENSG00000208308 |
. |
59324 |
0.14 |
| chr9_104203183_104203334 | 0.19 |
ALDOB |
aldolase B, fructose-bisphosphate |
5153 |
0.17 |
| chr11_6742912_6743421 | 0.19 |
GVINP1 |
GTPase, very large interferon inducible pseudogene 1 |
55 |
0.96 |
| chr17_42761038_42761322 | 0.19 |
CCDC43 |
coiled-coil domain containing 43 |
5913 |
0.13 |
| chr14_65173299_65173599 | 0.19 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
2134 |
0.36 |
| chr10_15713490_15713641 | 0.19 |
ITGA8 |
integrin, alpha 8 |
48559 |
0.18 |
| chr13_100138979_100139130 | 0.19 |
LINC00449 |
long intergenic non-protein coding RNA 449 |
14252 |
0.16 |
| chr2_135165435_135165594 | 0.19 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
153684 |
0.04 |
| chr1_175158282_175158496 | 0.19 |
KIAA0040 |
KIAA0040 |
3501 |
0.33 |
| chr20_23142717_23142901 | 0.19 |
ENSG00000201527 |
. |
1201 |
0.54 |
| chr3_5054013_5054242 | 0.19 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
32481 |
0.16 |
| chr10_75305750_75305901 | 0.19 |
ENSG00000207327 |
. |
16910 |
0.12 |
| chr14_74365658_74365809 | 0.19 |
ZNF410 |
zinc finger protein 410 |
4670 |
0.11 |
| chr22_35725194_35725345 | 0.19 |
ENSG00000266320 |
. |
6364 |
0.17 |
| chr1_229161136_229161564 | 0.19 |
RP5-1061H20.5 |
|
201959 |
0.02 |
| chr6_108958559_108958710 | 0.19 |
FOXO3 |
forkhead box O3 |
18915 |
0.27 |
| chr2_69006771_69007110 | 0.18 |
ARHGAP25 |
Rho GTPase activating protein 25 |
4868 |
0.27 |
| chr18_74138582_74138815 | 0.18 |
ZNF516 |
zinc finger protein 516 |
47830 |
0.13 |
| chr11_75488134_75488285 | 0.18 |
CTD-2530H12.2 |
|
1260 |
0.29 |
| chr8_20056663_20056818 | 0.18 |
ATP6V1B2 |
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
1790 |
0.34 |
| chr4_80499592_80499743 | 0.18 |
OR7E94P |
olfactory receptor, family 7, subfamily E, member 94 pseudogene |
9617 |
0.31 |
| chr3_151081140_151081291 | 0.18 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
21385 |
0.16 |
| chr9_114661606_114661757 | 0.18 |
UGCG |
UDP-glucose ceramide glucosyltransferase |
2635 |
0.29 |
| chr1_226223326_226223530 | 0.18 |
H3F3A |
H3 histone, family 3A |
26124 |
0.13 |
| chr2_228354322_228354473 | 0.18 |
AGFG1 |
ArfGAP with FG repeats 1 |
17309 |
0.19 |
| chr6_7312394_7312749 | 0.18 |
SSR1 |
signal sequence receptor, alpha |
836 |
0.65 |
| chr10_94002812_94003219 | 0.18 |
CPEB3 |
cytoplasmic polyadenylation element binding protein 3 |
12 |
0.98 |
| chr11_63274107_63274394 | 0.18 |
LGALS12 |
lectin, galactoside-binding, soluble, 12 |
415 |
0.81 |
| chr8_141114736_141114887 | 0.18 |
C8orf17 |
chromosome 8 open reading frame 17 |
171395 |
0.04 |
| chr5_1503133_1503343 | 0.18 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
20854 |
0.2 |
| chr1_155204670_155204827 | 0.18 |
ENSG00000216109 |
. |
1880 |
0.12 |
| chr20_56878561_56878712 | 0.18 |
PPP4R1L |
protein phosphatase 4, regulatory subunit 1-like |
5859 |
0.21 |
| chr4_185182646_185182797 | 0.18 |
ENSG00000221523 |
. |
5863 |
0.22 |
| chr2_7139293_7139444 | 0.18 |
RNF144A |
ring finger protein 144A |
2297 |
0.38 |
| chr15_91406561_91406712 | 0.18 |
FURIN |
furin (paired basic amino acid cleaving enzyme) |
5186 |
0.12 |
| chr6_152269247_152269414 | 0.18 |
ESR1 |
estrogen receptor 1 |
67538 |
0.13 |
| chr7_3786961_3787112 | 0.18 |
AC011284.3 |
|
103620 |
0.08 |
| chr2_216997149_216997386 | 0.18 |
XRCC5 |
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
23193 |
0.18 |
| chrX_10087005_10087156 | 0.18 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
37944 |
0.15 |
| chr8_82065263_82065697 | 0.18 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
41177 |
0.19 |
| chr16_23877339_23877739 | 0.18 |
PRKCB |
protein kinase C, beta |
28995 |
0.21 |
| chr8_61609604_61609755 | 0.18 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
17556 |
0.26 |
| chr3_43305774_43305925 | 0.18 |
SNRK |
SNF related kinase |
22155 |
0.18 |
| chr10_134068535_134068686 | 0.18 |
STK32C |
serine/threonine kinase 32C |
52108 |
0.12 |
| chr13_53601964_53602115 | 0.18 |
OLFM4 |
olfactomedin 4 |
855 |
0.75 |
| chr3_17040308_17040567 | 0.18 |
PLCL2 |
phospholipase C-like 2 |
11549 |
0.24 |
| chr1_225506705_225506856 | 0.18 |
DNAH14 |
dynein, axonemal, heavy chain 14 |
68415 |
0.11 |
| chr17_73965690_73965841 | 0.18 |
ACOX1 |
acyl-CoA oxidase 1, palmitoyl |
9009 |
0.09 |
| chr19_51613185_51613336 | 0.18 |
CTU1 |
cytosolic thiouridylase subunit 1 |
1633 |
0.2 |
| chr9_7966540_7966691 | 0.18 |
TMEM261 |
transmembrane protein 261 |
166548 |
0.04 |
| chr5_96157488_96157872 | 0.18 |
ERAP1 |
endoplasmic reticulum aminopeptidase 1 |
13877 |
0.14 |
| chr2_202322766_202322917 | 0.17 |
STRADB |
STE20-related kinase adaptor beta |
6322 |
0.19 |
| chr10_11249572_11249747 | 0.17 |
RP3-323N1.2 |
|
36320 |
0.17 |
| chr22_19399369_19399520 | 0.17 |
HIRA |
histone cell cycle regulator |
19775 |
0.1 |
| chr19_41913601_41913801 | 0.17 |
BCKDHA |
branched chain keto acid dehydrogenase E1, alpha polypeptide |
2926 |
0.12 |
| chr2_103037979_103038143 | 0.17 |
IL18RAP |
interleukin 18 receptor accessory protein |
1915 |
0.26 |
| chr21_16579711_16579862 | 0.17 |
NRIP1 |
nuclear receptor interacting protein 1 |
142465 |
0.05 |
| chr2_85901507_85901658 | 0.17 |
SFTPB |
surfactant protein B |
5718 |
0.14 |
| chr19_43086596_43086747 | 0.17 |
CEACAM8 |
carcinoembryonic antigen-related cell adhesion molecule 8 |
12411 |
0.18 |
| chr21_34792697_34792848 | 0.17 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
11854 |
0.16 |
| chr4_15666458_15666609 | 0.17 |
FBXL5 |
F-box and leucine-rich repeat protein 5 |
5043 |
0.21 |
| chr9_126440722_126440880 | 0.17 |
DENND1A |
DENN/MADD domain containing 1A |
26471 |
0.21 |
| chr8_123957813_123957964 | 0.17 |
RP11-557C18.3 |
|
56912 |
0.1 |
| chr7_37748458_37748609 | 0.17 |
GPR141 |
G protein-coupled receptor 141 |
25058 |
0.2 |
| chr19_42702815_42703281 | 0.17 |
ENSG00000265122 |
. |
5818 |
0.1 |
| chr4_13622958_13623109 | 0.17 |
BOD1L1 |
biorientation of chromosomes in cell division 1-like 1 |
6314 |
0.21 |
| chr16_68923648_68923799 | 0.17 |
ENSG00000199979 |
. |
4143 |
0.21 |
| chr7_42947190_42947341 | 0.17 |
C7orf25 |
chromosome 7 open reading frame 25 |
4244 |
0.24 |
| chr14_102319181_102319389 | 0.17 |
CTD-2017C7.1 |
|
13417 |
0.17 |
| chr21_15916640_15916923 | 0.17 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
1881 |
0.42 |
| chr10_30021965_30022179 | 0.17 |
SVIL |
supervillin |
2658 |
0.33 |
| chr1_109676026_109676177 | 0.17 |
KIAA1324 |
KIAA1324 |
19324 |
0.11 |
| chr2_136765754_136765905 | 0.17 |
AC093391.2 |
|
644 |
0.76 |
| chr1_6069612_6069763 | 0.17 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
16673 |
0.16 |
| chr20_46354413_46354564 | 0.17 |
CTD-2653D5.1 |
|
47843 |
0.14 |
| chr4_6687814_6687965 | 0.17 |
AC093323.1 |
Uncharacterized protein |
6300 |
0.15 |
| chr2_109883164_109883315 | 0.17 |
ENSG00000265965 |
. |
46842 |
0.18 |
| chr8_48422326_48422477 | 0.17 |
RP11-697N18.4 |
|
17558 |
0.25 |
| chr4_81064069_81064253 | 0.17 |
PRDM8 |
PR domain containing 8 |
41278 |
0.16 |
| chr11_27379616_27379767 | 0.17 |
CCDC34 |
coiled-coil domain containing 34 |
5090 |
0.29 |
| chr16_84377709_84377860 | 0.17 |
ATP2C2 |
ATPase, Ca++ transporting, type 2C, member 2 |
24349 |
0.15 |
| chr11_73131309_73131460 | 0.17 |
RP11-809N8.4 |
|
15042 |
0.16 |
| chr10_6436483_6436634 | 0.17 |
DKFZP667F0711 |
|
44280 |
0.18 |
| chr5_98276963_98277114 | 0.17 |
ENSG00000200351 |
. |
4587 |
0.25 |
| chr15_72660549_72660700 | 0.17 |
HEXA |
hexosaminidase A (alpha polypeptide) |
7561 |
0.16 |
| chr2_231074797_231075232 | 0.17 |
SP140 |
SP140 nuclear body protein |
7188 |
0.19 |
| chr2_43643825_43643976 | 0.17 |
ENSG00000252804 |
. |
8454 |
0.29 |
| chr17_53841712_53842007 | 0.17 |
PCTP |
phosphatidylcholine transfer protein |
9238 |
0.26 |
| chr5_111463867_111464018 | 0.17 |
ENSG00000238363 |
. |
33240 |
0.17 |
| chr14_65460947_65461098 | 0.16 |
FNTB |
farnesyltransferase, CAAX box, beta |
7584 |
0.12 |
| chr10_3506996_3507147 | 0.16 |
RP11-184A2.3 |
|
286188 |
0.01 |
| chr14_55583593_55583744 | 0.16 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
7160 |
0.23 |
| chr5_42631476_42631627 | 0.16 |
GHR |
growth hormone receptor |
65593 |
0.13 |
| chrX_30657144_30657355 | 0.16 |
GK |
glycerol kinase |
14227 |
0.19 |
| chr16_81962423_81962574 | 0.16 |
ENSG00000260682 |
. |
33033 |
0.17 |
| chr13_99931097_99931381 | 0.16 |
GPR18 |
G protein-coupled receptor 18 |
17241 |
0.18 |
| chr12_104192450_104192601 | 0.16 |
RP11-650K20.2 |
|
20863 |
0.13 |
| chr16_53534714_53534865 | 0.16 |
AKTIP |
AKT interacting protein |
548 |
0.79 |
| chr12_95042694_95042845 | 0.16 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
1569 |
0.51 |
| chr16_3867727_3867878 | 0.16 |
CREBBP |
CREB binding protein |
36934 |
0.16 |
| chr17_45995658_45995845 | 0.16 |
RP11-6N17.3 |
|
14721 |
0.09 |
| chr15_68507516_68507667 | 0.16 |
CALML4 |
calmodulin-like 4 |
9174 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.2 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0031034 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.1 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0002448 | mast cell mediated immunity(GO:0002448) |
| 0.0 | 0.1 | GO:0021779 | oligodendrocyte cell fate commitment(GO:0021779) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0046449 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.0 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.0 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.0 | GO:0060751 | branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.2 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.0 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0060192 | negative regulation of lipoprotein lipase activity(GO:0051005) negative regulation of lipase activity(GO:0060192) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0030540 | female genitalia development(GO:0030540) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.0 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0034442 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 | 0.0 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.0 | GO:0017000 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0043032 | positive regulation of macrophage activation(GO:0043032) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.0 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.0 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.0 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0060678 | dichotomous subdivision of terminal units involved in ureteric bud branching(GO:0060678) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.0 | GO:1903504 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0032375 | negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) |
| 0.0 | 0.0 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0042891 | tetracycline transport(GO:0015904) antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.1 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0030354 | melanin-concentrating hormone activity(GO:0030354) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0031420 | alkali metal ion binding(GO:0031420) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |