Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for TBX20
Z-value: 0.63
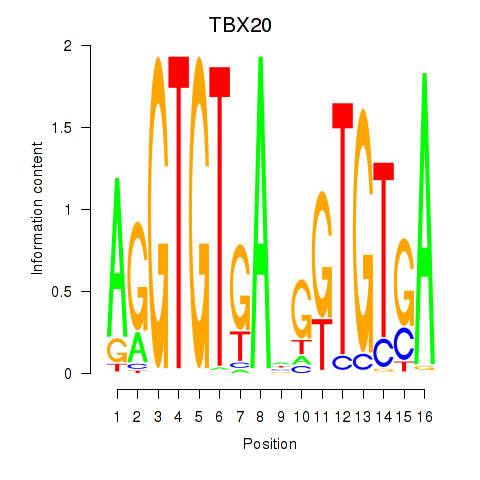
Transcription factors associated with TBX20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX20
|
ENSG00000164532.10 | T-box transcription factor 20 |
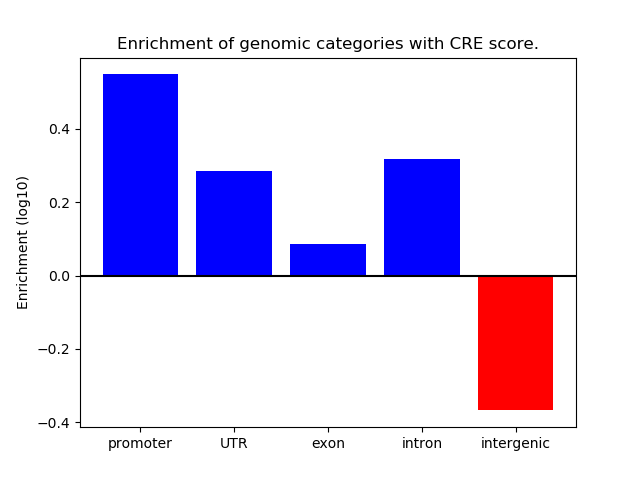
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_35293912_35294283 | TBX20 | 339 | 0.929839 | 0.72 | 2.7e-02 | Click! |
| chr7_35420213_35420364 | TBX20 | 126530 | 0.057190 | 0.53 | 1.5e-01 | Click! |
| chr7_35291561_35291712 | TBX20 | 2122 | 0.437799 | 0.40 | 2.9e-01 | Click! |
| chr7_35297544_35297695 | TBX20 | 3861 | 0.340226 | 0.37 | 3.3e-01 | Click! |
| chr7_35293101_35293402 | TBX20 | 507 | 0.870114 | 0.30 | 4.3e-01 | Click! |
Activity of the TBX20 motif across conditions
Conditions sorted by the z-value of the TBX20 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
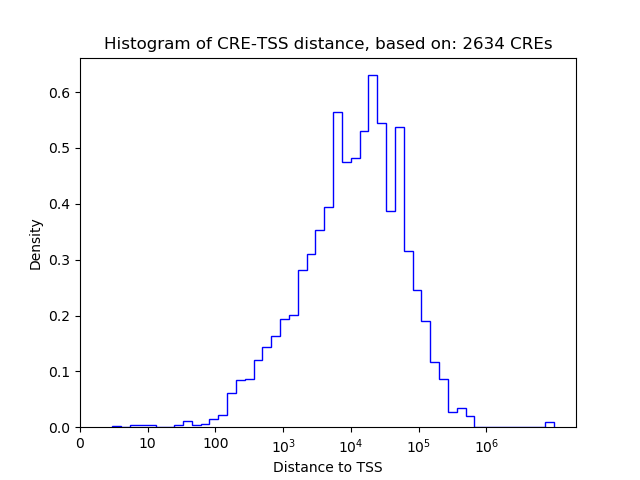
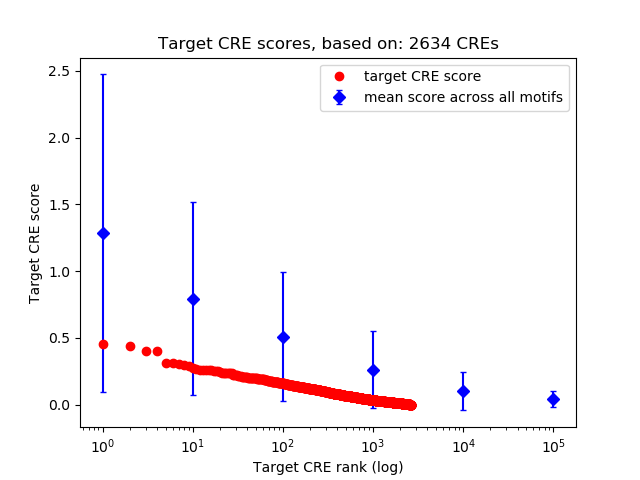
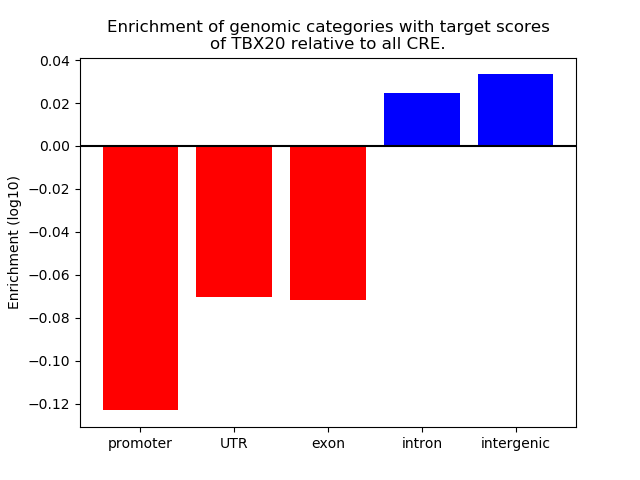
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_101173362_101173513 | 0.45 |
DLK1 |
delta-like 1 homolog (Drosophila) |
18605 |
0.11 |
| chr12_4353084_4353235 | 0.44 |
CCND2 |
cyclin D2 |
29779 |
0.15 |
| chr17_7012818_7013162 | 0.41 |
ASGR2 |
asialoglycoprotein receptor 2 |
4569 |
0.12 |
| chr11_118762748_118763140 | 0.40 |
RP11-158I9.5 |
|
4493 |
0.11 |
| chr11_1802177_1802346 | 0.31 |
CTSD |
cathepsin D |
17039 |
0.08 |
| chr8_96228209_96228362 | 0.31 |
C8orf37 |
chromosome 8 open reading frame 37 |
53144 |
0.14 |
| chr16_81866339_81866536 | 0.30 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
50437 |
0.16 |
| chr18_74086568_74086771 | 0.30 |
RP11-504I13.3 |
|
1997 |
0.35 |
| chr5_10644618_10644885 | 0.29 |
ANKRD33B-AS1 |
ANKRD33B antisense RNA 1 |
16414 |
0.23 |
| chr18_60917029_60917180 | 0.27 |
ENSG00000238988 |
. |
55206 |
0.11 |
| chr9_92755952_92756103 | 0.27 |
ENSG00000263967 |
. |
29790 |
0.27 |
| chr5_154164340_154164572 | 0.26 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
7837 |
0.19 |
| chr12_65045752_65045922 | 0.26 |
RP11-338E21.3 |
|
2962 |
0.18 |
| chr14_22562409_22562689 | 0.26 |
ENSG00000238634 |
. |
48338 |
0.17 |
| chr12_68025791_68026092 | 0.26 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
16177 |
0.25 |
| chr14_66304826_66305118 | 0.26 |
CTD-2014B16.3 |
Uncharacterized protein |
166269 |
0.04 |
| chr3_45906834_45907101 | 0.25 |
CCR9 |
chemokine (C-C motif) receptor 9 |
21029 |
0.15 |
| chr10_47638607_47638777 | 0.25 |
ANTXRLP1 |
anthrax toxin receptor-like pseudogene 1 |
6099 |
0.23 |
| chr18_33389231_33389398 | 0.25 |
ENSG00000207797 |
. |
95575 |
0.08 |
| chr20_56201390_56201672 | 0.25 |
ZBP1 |
Z-DNA binding protein 1 |
5899 |
0.25 |
| chr13_46743886_46744372 | 0.24 |
ENSG00000240767 |
. |
246 |
0.88 |
| chr3_43813256_43813407 | 0.24 |
ABHD5 |
abhydrolase domain containing 5 |
60132 |
0.14 |
| chr4_140973396_140973608 | 0.24 |
RP11-392B6.1 |
|
75667 |
0.11 |
| chr2_68615070_68615679 | 0.24 |
AC015969.3 |
|
22658 |
0.16 |
| chr1_206942223_206942745 | 0.24 |
IL10 |
interleukin 10 |
3355 |
0.2 |
| chr11_128589269_128589420 | 0.24 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23426 |
0.17 |
| chr2_198079971_198080247 | 0.23 |
ANKRD44 |
ankyrin repeat domain 44 |
17347 |
0.19 |
| chr3_37499337_37499488 | 0.23 |
ITGA9 |
integrin, alpha 9 |
5802 |
0.25 |
| chr2_106426708_106426906 | 0.22 |
NCK2 |
NCK adaptor protein 2 |
6208 |
0.28 |
| chr10_11636550_11636701 | 0.22 |
RP11-138I18.1 |
|
16679 |
0.22 |
| chr19_54789021_54789175 | 0.22 |
ENSG00000264703 |
. |
3134 |
0.11 |
| chr8_581194_581388 | 0.22 |
ERICH1 |
glutamate-rich 1 |
42366 |
0.17 |
| chr15_101779146_101779303 | 0.21 |
CHSY1 |
chondroitin sulfate synthase 1 |
12913 |
0.18 |
| chr17_8848631_8848849 | 0.21 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
20284 |
0.21 |
| chr6_24924284_24924680 | 0.21 |
FAM65B |
family with sequence similarity 65, member B |
11706 |
0.23 |
| chr15_72663552_72663703 | 0.21 |
HEXA |
hexosaminidase A (alpha polypeptide) |
4558 |
0.17 |
| chr18_60774830_60775182 | 0.21 |
RP11-299P2.1 |
|
43547 |
0.17 |
| chr13_30947293_30948038 | 0.21 |
KATNAL1 |
katanin p60 subunit A-like 1 |
66044 |
0.12 |
| chr6_13688959_13689110 | 0.21 |
RANBP9 |
RAN binding protein 9 |
1854 |
0.32 |
| chr16_84121409_84121596 | 0.21 |
MBTPS1 |
membrane-bound transcription factor peptidase, site 1 |
28898 |
0.11 |
| chr22_19607046_19607250 | 0.20 |
CLDN5 |
claudin 5 |
92080 |
0.06 |
| chr9_134019215_134019366 | 0.20 |
NUP214 |
nucleoporin 214kDa |
7915 |
0.15 |
| chr6_30475738_30475914 | 0.20 |
XXbac-BPG249D20.9 |
|
8217 |
0.13 |
| chr3_43952236_43952387 | 0.20 |
ENSG00000252980 |
. |
160268 |
0.04 |
| chr2_182048541_182048785 | 0.20 |
ENSG00000266705 |
. |
121716 |
0.06 |
| chr2_96920389_96920540 | 0.20 |
TMEM127 |
transmembrane protein 127 |
5869 |
0.15 |
| chr1_64708785_64708944 | 0.20 |
UBE2U |
ubiquitin-conjugating enzyme E2U (putative) |
22317 |
0.23 |
| chr21_16045329_16045670 | 0.20 |
AF165138.7 |
Protein LOC388813 |
14357 |
0.26 |
| chr11_126206744_126207042 | 0.20 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
18662 |
0.11 |
| chr4_2787896_2788215 | 0.20 |
SH3BP2 |
SH3-domain binding protein 2 |
6695 |
0.21 |
| chr3_53174315_53174466 | 0.20 |
RFT1 |
RFT1 homolog (S. cerevisiae) |
9912 |
0.17 |
| chr12_7811014_7811165 | 0.20 |
APOBEC1 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
7400 |
0.18 |
| chr8_1803402_1803553 | 0.19 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
27345 |
0.19 |
| chr11_111820278_111820429 | 0.19 |
DIXDC1 |
DIX domain containing 1 |
12202 |
0.11 |
| chr13_114814991_114815196 | 0.19 |
RASA3 |
RAS p21 protein activator 3 |
28345 |
0.21 |
| chr3_131221293_131221589 | 0.19 |
MRPL3 |
mitochondrial ribosomal protein L3 |
340 |
0.87 |
| chr6_6912917_6913068 | 0.19 |
ENSG00000240936 |
. |
26138 |
0.25 |
| chr16_11325488_11325654 | 0.19 |
RMI2 |
RecQ mediated genome instability 2 |
17935 |
0.1 |
| chr19_12892440_12892622 | 0.19 |
HOOK2 |
hook microtubule-tethering protein 2 |
3260 |
0.09 |
| chr20_47417901_47418129 | 0.19 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
26405 |
0.22 |
| chr14_91718270_91718619 | 0.19 |
GPR68 |
G protein-coupled receptor 68 |
1780 |
0.32 |
| chr17_3864337_3864743 | 0.19 |
ATP2A3 |
ATPase, Ca++ transporting, ubiquitous |
3045 |
0.24 |
| chr4_2293050_2293361 | 0.18 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
3763 |
0.15 |
| chr9_35834312_35834712 | 0.18 |
TMEM8B |
transmembrane protein 8B |
4999 |
0.1 |
| chr4_152586514_152586670 | 0.18 |
RP11-164P12.4 |
|
1784 |
0.38 |
| chr20_17764632_17764807 | 0.18 |
ENSG00000221220 |
. |
58508 |
0.11 |
| chrX_18686644_18686795 | 0.18 |
RS1 |
retinoschisin 1 |
3510 |
0.25 |
| chr2_172864951_172865102 | 0.18 |
METAP1D |
methionyl aminopeptidase type 1D (mitochondrial) |
536 |
0.83 |
| chr12_65061545_65061862 | 0.18 |
RP11-338E21.3 |
|
12904 |
0.13 |
| chr3_121794531_121794682 | 0.18 |
CD86 |
CD86 molecule |
2098 |
0.34 |
| chr2_191514617_191514768 | 0.18 |
NAB1 |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
696 |
0.73 |
| chr2_99076206_99076588 | 0.18 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
14984 |
0.23 |
| chr2_97590837_97591148 | 0.18 |
FAM178B |
family with sequence similarity 178, member B |
27122 |
0.12 |
| chrX_40240519_40240688 | 0.18 |
ENSG00000238920 |
. |
96254 |
0.08 |
| chr6_111180825_111181074 | 0.17 |
ENSG00000199360 |
. |
3329 |
0.2 |
| chr17_76129423_76129727 | 0.17 |
TMC6 |
transmembrane channel-like 6 |
1087 |
0.35 |
| chr19_7192448_7192688 | 0.17 |
INSR |
insulin receptor |
24579 |
0.16 |
| chr6_166673464_166673615 | 0.17 |
PRR18 |
proline rich 18 |
48332 |
0.13 |
| chr7_37711541_37711746 | 0.17 |
GPR141 |
G protein-coupled receptor 141 |
11757 |
0.23 |
| chr11_73694860_73695069 | 0.17 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
612 |
0.66 |
| chr17_56419519_56419979 | 0.17 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
5030 |
0.12 |
| chr2_26678199_26678350 | 0.17 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
7107 |
0.21 |
| chr13_30888906_30889057 | 0.17 |
KATNAL1 |
katanin p60 subunit A-like 1 |
7360 |
0.26 |
| chr14_70092927_70093173 | 0.17 |
KIAA0247 |
KIAA0247 |
14737 |
0.23 |
| chr10_10924376_10924527 | 0.17 |
CELF2 |
CUGBP, Elav-like family member 2 |
122808 |
0.06 |
| chr2_144179957_144180310 | 0.17 |
AC096558.1 |
|
58217 |
0.15 |
| chr10_111897699_111897850 | 0.17 |
MXI1 |
MAX interactor 1, dimerization protein |
69589 |
0.1 |
| chr4_185373165_185373459 | 0.17 |
ENSG00000251878 |
. |
20192 |
0.17 |
| chr20_827974_828208 | 0.17 |
FAM110A |
family with sequence similarity 110, member A |
2324 |
0.35 |
| chr3_18531533_18531684 | 0.17 |
ENSG00000228956 |
. |
24124 |
0.22 |
| chr9_80516027_80516178 | 0.17 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
78187 |
0.12 |
| chr6_17451609_17451760 | 0.17 |
CAP2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
57741 |
0.15 |
| chr1_235806441_235806592 | 0.16 |
GNG4 |
guanine nucleotide binding protein (G protein), gamma 4 |
6777 |
0.28 |
| chr1_27951892_27952043 | 0.16 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
679 |
0.61 |
| chr5_6632202_6632695 | 0.16 |
NSUN2 |
NOP2/Sun RNA methyltransferase family, member 2 |
707 |
0.55 |
| chr1_204452670_204453343 | 0.16 |
PIK3C2B |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
6440 |
0.2 |
| chr4_48212801_48212952 | 0.16 |
ENSG00000202014 |
. |
57425 |
0.11 |
| chr3_5222978_5223174 | 0.16 |
AC026202.3 |
|
5938 |
0.16 |
| chr3_49370394_49370545 | 0.16 |
USP4 |
ubiquitin specific peptidase 4 (proto-oncogene) |
6997 |
0.12 |
| chr12_2753054_2753205 | 0.16 |
CACNA1C-AS2 |
CACNA1C antisense RNA 2 |
28257 |
0.16 |
| chr3_122090075_122090234 | 0.16 |
CCDC58 |
coiled-coil domain containing 58 |
1715 |
0.31 |
| chr3_18028540_18028691 | 0.16 |
TBC1D5 |
TBC1 domain family, member 5 |
244471 |
0.02 |
| chr9_29618271_29618422 | 0.16 |
ENSG00000239155 |
. |
560229 |
0.0 |
| chr3_57657383_57657934 | 0.16 |
DENND6A |
DENN/MADD domain containing 6A |
11105 |
0.15 |
| chr16_12549529_12549680 | 0.16 |
RP11-165M1.2 |
|
82924 |
0.07 |
| chr14_102667759_102667910 | 0.16 |
WDR20 |
WD repeat domain 20 |
6551 |
0.2 |
| chr8_134925670_134925821 | 0.16 |
ENSG00000212273 |
. |
268777 |
0.02 |
| chr1_209778895_209779046 | 0.16 |
LAMB3 |
laminin, beta 3 |
13173 |
0.16 |
| chr10_111978388_111978584 | 0.15 |
MXI1 |
MAX interactor 1, dimerization protein |
7265 |
0.22 |
| chr10_22938945_22939188 | 0.15 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
58424 |
0.15 |
| chr12_48336587_48336738 | 0.15 |
VDR |
vitamin D (1,25- dihydroxyvitamin D3) receptor |
169 |
0.94 |
| chr16_88546371_88546522 | 0.15 |
ENSG00000263456 |
. |
11120 |
0.16 |
| chr19_6663451_6663602 | 0.15 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
6602 |
0.15 |
| chr2_100625887_100626113 | 0.15 |
AFF3 |
AF4/FMR2 family, member 3 |
94992 |
0.08 |
| chr8_95975747_95976108 | 0.15 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
5691 |
0.16 |
| chr17_13766234_13766385 | 0.15 |
ENSG00000236088 |
. |
29847 |
0.25 |
| chr5_35794795_35794959 | 0.15 |
SPEF2 |
sperm flagellar 2 |
15607 |
0.2 |
| chr1_29252967_29253119 | 0.15 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
11952 |
0.18 |
| chr7_2292947_2293098 | 0.15 |
NUDT1 |
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
10482 |
0.17 |
| chr12_93577696_93578039 | 0.15 |
RP11-511B23.2 |
|
31583 |
0.17 |
| chr16_15929944_15930095 | 0.15 |
MYH11 |
myosin, heavy chain 11, smooth muscle |
20849 |
0.15 |
| chr8_41350711_41350862 | 0.15 |
GOLGA7 |
golgin A7 |
2613 |
0.25 |
| chr22_43349779_43349930 | 0.15 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
6030 |
0.25 |
| chr2_68994501_68994719 | 0.15 |
ARHGAP25 |
Rho GTPase activating protein 25 |
7323 |
0.25 |
| chr20_10455725_10456172 | 0.15 |
SLX4IP |
SLX4 interacting protein |
39997 |
0.16 |
| chr7_128500206_128500480 | 0.15 |
RP11-309L24.2 |
|
2337 |
0.2 |
| chr12_90272594_90272745 | 0.15 |
ENSG00000252823 |
. |
124833 |
0.06 |
| chr2_60376405_60376556 | 0.14 |
ENSG00000200807 |
. |
235260 |
0.02 |
| chr4_68410846_68410997 | 0.14 |
CENPC |
centromere protein C |
403 |
0.87 |
| chr4_102203245_102203491 | 0.14 |
ENSG00000221265 |
. |
48203 |
0.15 |
| chr9_101797611_101797762 | 0.14 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
68634 |
0.1 |
| chr13_46507098_46507337 | 0.14 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
36588 |
0.18 |
| chr7_99818571_99819087 | 0.14 |
ENSG00000222482 |
. |
1086 |
0.28 |
| chr12_9768783_9768954 | 0.14 |
ENSG00000212345 |
. |
7956 |
0.13 |
| chr22_43324537_43324688 | 0.14 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
17267 |
0.21 |
| chr9_37196104_37196255 | 0.14 |
ENSG00000200502 |
. |
36045 |
0.16 |
| chr22_40783922_40784073 | 0.14 |
RP5-1042K10.10 |
|
224 |
0.92 |
| chr15_70851843_70852459 | 0.14 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
142469 |
0.05 |
| chr11_118186120_118186587 | 0.14 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
10739 |
0.12 |
| chr2_112912774_112913193 | 0.14 |
FBLN7 |
fibulin 7 |
4342 |
0.28 |
| chr14_89864185_89864473 | 0.14 |
RP11-33N16.2 |
|
2010 |
0.27 |
| chr20_48276557_48276708 | 0.14 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
53783 |
0.11 |
| chr4_166177052_166177792 | 0.14 |
ENSG00000200974 |
. |
3690 |
0.24 |
| chr16_72958519_72958670 | 0.14 |
ENSG00000221799 |
. |
60162 |
0.12 |
| chr1_179675259_179675410 | 0.14 |
ENSG00000221547 |
. |
4147 |
0.23 |
| chr2_99069791_99070349 | 0.14 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
8657 |
0.24 |
| chr13_74318994_74319145 | 0.14 |
KLF12 |
Kruppel-like factor 12 |
250117 |
0.02 |
| chr19_36704820_36705397 | 0.14 |
ZNF146 |
zinc finger protein 146 |
396 |
0.49 |
| chr5_10448843_10449058 | 0.14 |
ROPN1L |
rhophilin associated tail protein 1-like |
6962 |
0.15 |
| chr6_111855590_111855741 | 0.14 |
TRAF3IP2-AS1 |
TRAF3IP2 antisense RNA 1 |
30789 |
0.15 |
| chr17_32281966_32282117 | 0.14 |
RP11-17M24.1 |
|
18816 |
0.19 |
| chr7_36199399_36199550 | 0.14 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
6597 |
0.24 |
| chr22_48533700_48533851 | 0.14 |
ENSG00000266508 |
. |
136401 |
0.05 |
| chr1_65241133_65241333 | 0.14 |
RAVER2 |
ribonucleoprotein, PTB-binding 2 |
2514 |
0.42 |
| chr20_61272668_61272952 | 0.13 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
987 |
0.37 |
| chr4_14272910_14273061 | 0.13 |
ENSG00000202449 |
. |
419349 |
0.01 |
| chr2_131138043_131138333 | 0.13 |
PTPN18 |
protein tyrosine phosphatase, non-receptor type 18 (brain-derived) |
10526 |
0.15 |
| chr12_10222553_10222837 | 0.13 |
ENSG00000223042 |
. |
14090 |
0.12 |
| chr5_118694501_118694652 | 0.13 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
2846 |
0.3 |
| chr11_1339801_1339952 | 0.13 |
TOLLIP-AS1 |
TOLLIP antisense RNA 1 (head to head) |
8877 |
0.15 |
| chr14_25162357_25162508 | 0.13 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
58959 |
0.11 |
| chr3_9823163_9823342 | 0.13 |
ARPC4 |
actin related protein 2/3 complex, subunit 4, 20kDa |
10927 |
0.09 |
| chr9_139260248_139260450 | 0.13 |
DNLZ |
DNL-type zinc finger |
2108 |
0.19 |
| chr10_114134943_114135530 | 0.13 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
213 |
0.94 |
| chrX_1388495_1388646 | 0.13 |
CSF2RA |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
832 |
0.46 |
| chrY_1338495_1338646 | 0.13 |
NA |
NA |
> 106 |
NA |
| chr1_159797270_159797554 | 0.13 |
SLAMF8 |
SLAM family member 8 |
811 |
0.47 |
| chrX_67917027_67917470 | 0.13 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
3755 |
0.36 |
| chrX_41416950_41417163 | 0.13 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
3824 |
0.22 |
| chr17_33840627_33840778 | 0.13 |
RP11-1094M14.5 |
|
3413 |
0.16 |
| chr10_98424639_98424798 | 0.13 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
4650 |
0.24 |
| chr8_103135143_103135295 | 0.13 |
NCALD |
neurocalcin delta |
959 |
0.56 |
| chr12_122327188_122327339 | 0.13 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
571 |
0.69 |
| chr13_41052110_41052363 | 0.13 |
AL133318.1 |
Uncharacterized protein |
59087 |
0.14 |
| chr17_31994111_31994311 | 0.13 |
RP11-31I22.2 |
|
40165 |
0.18 |
| chr13_114271259_114271410 | 0.13 |
TFDP1 |
transcription factor Dp-1 |
31593 |
0.15 |
| chr2_87874778_87875151 | 0.13 |
ENSG00000265507 |
. |
54310 |
0.16 |
| chr3_39276331_39276557 | 0.13 |
XIRP1 |
xin actin-binding repeat containing 1 |
42357 |
0.13 |
| chr9_90169617_90169893 | 0.13 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
1386 |
0.54 |
| chr17_3876410_3876657 | 0.13 |
ATP2A3 |
ATPase, Ca++ transporting, ubiquitous |
8797 |
0.19 |
| chr1_77924703_77924854 | 0.12 |
ENSG00000251767 |
. |
38768 |
0.16 |
| chr3_47026416_47026785 | 0.12 |
CCDC12 |
coiled-coil domain containing 12 |
3128 |
0.23 |
| chr2_240962359_240962510 | 0.12 |
NDUFA10 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa |
2289 |
0.24 |
| chr6_73349801_73349952 | 0.12 |
KCNQ5-IT1 |
KCNQ5 intronic transcript 1 (non-protein coding) |
9653 |
0.25 |
| chr1_9778118_9778444 | 0.12 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
154 |
0.95 |
| chr6_107940361_107940512 | 0.12 |
RP1-67A8.3 |
|
107511 |
0.06 |
| chr5_66343612_66343763 | 0.12 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
43204 |
0.17 |
| chr2_31441668_31441819 | 0.12 |
CAPN14 |
calpain 14 |
1332 |
0.44 |
| chr13_114917830_114917981 | 0.12 |
RASA3 |
RAS p21 protein activator 3 |
19819 |
0.2 |
| chr7_26895297_26895448 | 0.12 |
SKAP2 |
src kinase associated phosphoprotein 2 |
1867 |
0.45 |
| chr2_109553737_109553904 | 0.12 |
EDAR |
ectodysplasin A receptor |
51905 |
0.16 |
| chr3_109106804_109106955 | 0.12 |
DPPA4 |
developmental pluripotency associated 4 |
50460 |
0.15 |
| chr10_64380278_64380429 | 0.12 |
ZNF365 |
zinc finger protein 365 |
23180 |
0.22 |
| chr17_72748432_72748583 | 0.12 |
MIR3615 |
microRNA 3615 |
3456 |
0.12 |
| chr13_52401708_52401859 | 0.12 |
RP11-327P2.5 |
|
23350 |
0.16 |
| chr13_21918498_21918871 | 0.12 |
ENSG00000222747 |
. |
25354 |
0.16 |
| chr2_231663134_231663369 | 0.12 |
ENSG00000201044 |
. |
46395 |
0.13 |
| chr20_22967144_22967295 | 0.12 |
RP4-753D10.5 |
|
44203 |
0.12 |
| chr3_138016758_138016909 | 0.12 |
NME9 |
NME/NM23 family member 9 |
21589 |
0.18 |
| chr11_116877018_116877229 | 0.12 |
ENSG00000264344 |
. |
9122 |
0.17 |
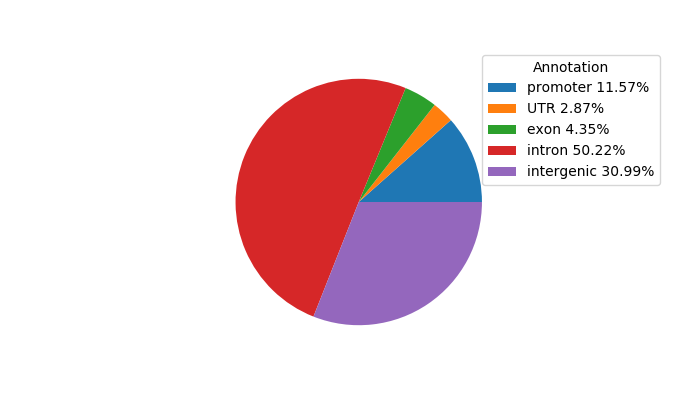
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.2 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.0 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0051006 | positive regulation of lipoprotein lipase activity(GO:0051006) positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.0 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |