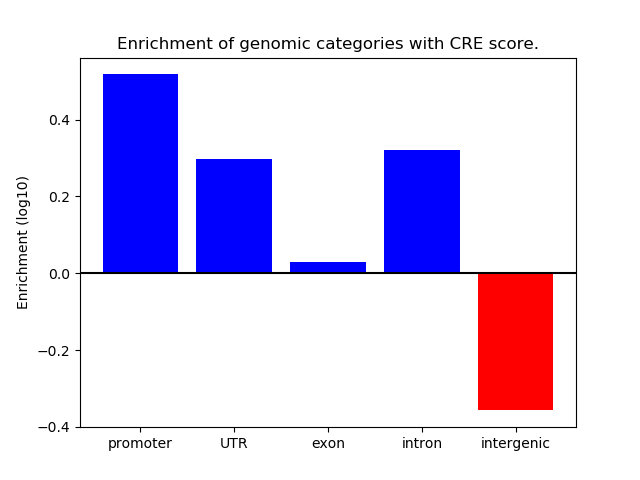
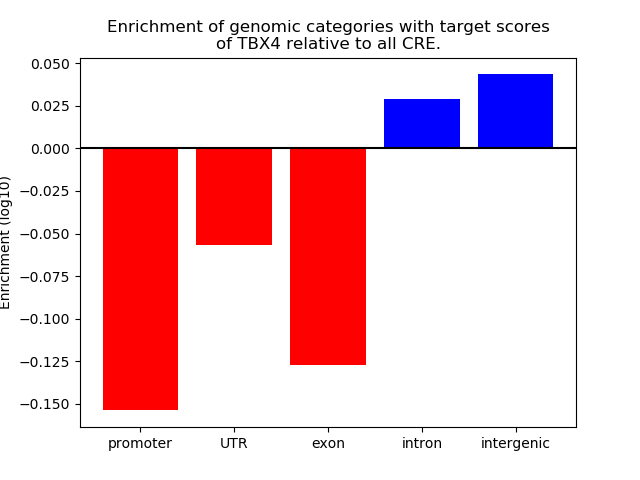
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
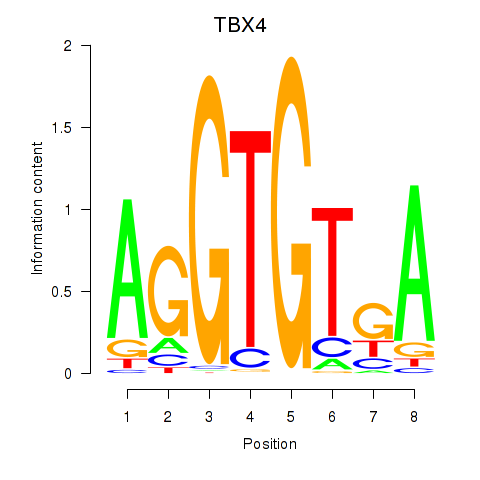
Results for TBX4
Z-value: 0.31
Transcription factors associated with TBX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX4
|
ENSG00000121075.5 | T-box transcription factor 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_59534235_59534493 | TBX4 | 557 | 0.716020 | -0.84 | 4.7e-03 | Click! |
| chr17_59534639_59534994 | TBX4 | 1009 | 0.480485 | -0.70 | 3.7e-02 | Click! |
| chr17_59535127_59535328 | TBX4 | 1420 | 0.357722 | -0.54 | 1.4e-01 | Click! |
| chr17_59529466_59529654 | TBX4 | 205 | 0.923279 | -0.53 | 1.4e-01 | Click! |
| chr17_59533558_59533709 | TBX4 | 174 | 0.937318 | -0.48 | 2.0e-01 | Click! |
Activity of the TBX4 motif across conditions
Conditions sorted by the z-value of the TBX4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
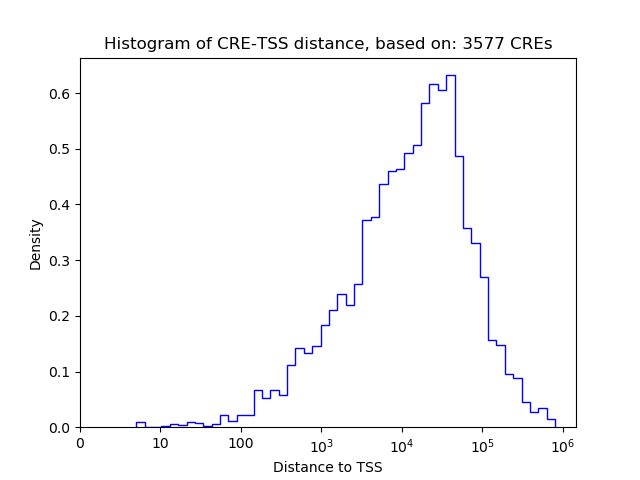
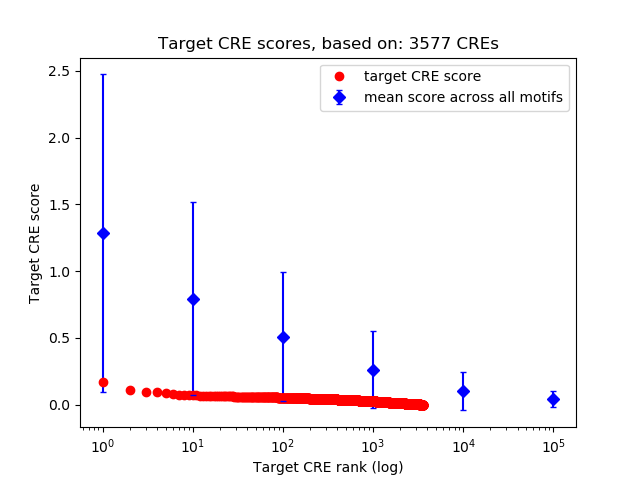
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_4584081_4584405 | 0.17 |
ENSG00000207124 |
. |
27099 |
0.26 |
| chr2_65065663_65065834 | 0.11 |
ENSG00000199964 |
. |
4457 |
0.21 |
| chr8_71515545_71515696 | 0.10 |
ENSG00000238450 |
. |
1147 |
0.39 |
| chr8_49228853_49229154 | 0.09 |
ENSG00000252710 |
. |
8413 |
0.32 |
| chr11_61448106_61448684 | 0.09 |
DAGLA |
diacylglycerol lipase, alpha |
490 |
0.8 |
| chr11_2117665_2117864 | 0.08 |
ENSG00000207805 |
. |
37675 |
0.09 |
| chr15_62737179_62737605 | 0.07 |
TLN2 |
talin 2 |
116172 |
0.06 |
| chr1_52435800_52436083 | 0.07 |
ENSG00000200839 |
. |
3141 |
0.18 |
| chr11_74742638_74742916 | 0.07 |
NEU3 |
sialidase 3 (membrane sialidase) |
42787 |
0.11 |
| chr9_91144416_91144843 | 0.07 |
NXNL2 |
nucleoredoxin-like 2 |
5387 |
0.32 |
| chr1_22253677_22253895 | 0.07 |
HSPG2 |
heparan sulfate proteoglycan 2 |
10004 |
0.15 |
| chr8_99170182_99170517 | 0.07 |
ENSG00000252558 |
. |
34192 |
0.13 |
| chr1_22252730_22253001 | 0.07 |
HSPG2 |
heparan sulfate proteoglycan 2 |
10925 |
0.15 |
| chr11_86458885_86459036 | 0.07 |
PRSS23 |
protease, serine, 23 |
43141 |
0.18 |
| chr20_4207508_4207659 | 0.07 |
ADRA1D |
adrenoceptor alpha 1D |
22138 |
0.19 |
| chr3_42078593_42078744 | 0.06 |
TRAK1 |
trafficking protein, kinesin binding 1 |
53894 |
0.14 |
| chr12_63139988_63140139 | 0.06 |
ENSG00000238475 |
. |
95108 |
0.07 |
| chr13_34931777_34932168 | 0.06 |
ENSG00000199196 |
. |
256997 |
0.02 |
| chr3_64172202_64172494 | 0.06 |
PRICKLE2-AS3 |
PRICKLE2 antisense RNA 3 |
872 |
0.63 |
| chr3_193988294_193988600 | 0.06 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
83600 |
0.08 |
| chr3_8580450_8580601 | 0.06 |
LMCD1 |
LIM and cysteine-rich domains 1 |
1656 |
0.43 |
| chr2_45581409_45581560 | 0.06 |
SRBD1 |
S1 RNA binding domain 1 |
213670 |
0.02 |
| chr5_178703659_178703833 | 0.06 |
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
68685 |
0.12 |
| chr19_42592250_42592591 | 0.06 |
POU2F2 |
POU class 2 homeobox 2 |
7137 |
0.11 |
| chr5_171288121_171288370 | 0.06 |
SMIM23 |
small integral membrane protein 23 |
75355 |
0.09 |
| chr3_159338407_159338558 | 0.06 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
143020 |
0.04 |
| chr5_64493902_64494516 | 0.06 |
ENSG00000207439 |
. |
75013 |
0.12 |
| chr14_105439124_105439578 | 0.06 |
AHNAK2 |
AHNAK nucleoprotein 2 |
5343 |
0.17 |
| chr2_217640205_217640356 | 0.06 |
ENSG00000222832 |
. |
18043 |
0.18 |
| chr11_44010465_44010739 | 0.06 |
C11orf96 |
chromosome 11 open reading frame 96 |
46547 |
0.11 |
| chrX_150958201_150958352 | 0.06 |
CNGA2 |
cyclic nucleotide gated channel alpha 2 |
51353 |
0.13 |
| chr10_64386237_64386388 | 0.06 |
ZNF365 |
zinc finger protein 365 |
17221 |
0.23 |
| chr16_29836315_29836998 | 0.06 |
MVP |
major vault protein |
4010 |
0.08 |
| chr1_66291138_66291289 | 0.06 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
32349 |
0.21 |
| chr7_36344154_36344305 | 0.06 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
7476 |
0.18 |
| chr15_34348811_34348962 | 0.06 |
AVEN |
apoptosis, caspase activation inhibitor |
17509 |
0.17 |
| chr1_240399314_240399591 | 0.06 |
FMN2 |
formin 2 |
9108 |
0.25 |
| chr4_169449939_169450239 | 0.06 |
PALLD |
palladin, cytoskeletal associated protein |
17370 |
0.21 |
| chr12_16119043_16119194 | 0.06 |
DERA |
deoxyribose-phosphate aldolase (putative) |
9596 |
0.27 |
| chr1_201666517_201666668 | 0.06 |
ENSG00000264802 |
. |
22044 |
0.12 |
| chr20_42161917_42162068 | 0.06 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
4645 |
0.17 |
| chr4_156726912_156727063 | 0.06 |
GUCY1B3 |
guanylate cyclase 1, soluble, beta 3 |
46376 |
0.13 |
| chr15_42213065_42213339 | 0.06 |
CTD-2382E5.4 |
|
413 |
0.76 |
| chr3_187800607_187800758 | 0.06 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
70390 |
0.11 |
| chr16_87615099_87615250 | 0.06 |
RP11-482M8.3 |
|
20622 |
0.17 |
| chr1_170675865_170676016 | 0.06 |
PRRX1 |
paired related homeobox 1 |
42862 |
0.19 |
| chr1_109045119_109045270 | 0.06 |
NBPF6 |
neuroblastoma breakpoint family, member 6 |
52251 |
0.14 |
| chr6_169497993_169498144 | 0.06 |
XXyac-YX65C7_A.2 |
|
115281 |
0.07 |
| chr2_20290862_20291052 | 0.06 |
RP11-644K8.1 |
|
39062 |
0.13 |
| chr13_50931215_50931366 | 0.06 |
ENSG00000221198 |
. |
6083 |
0.33 |
| chr14_100435801_100435952 | 0.06 |
EVL |
Enah/Vasp-like |
1910 |
0.34 |
| chr17_48281191_48281342 | 0.06 |
COL1A1 |
collagen, type I, alpha 1 |
2273 |
0.19 |
| chr1_235044220_235044371 | 0.06 |
ENSG00000239690 |
. |
4362 |
0.31 |
| chr3_11739592_11739743 | 0.06 |
VGLL4 |
vestigial like 4 (Drosophila) |
12344 |
0.26 |
| chr20_48768588_48769651 | 0.06 |
TMEM189-UBE2V1 |
TMEM189-UBE2V1 readthrough |
1055 |
0.37 |
| chr11_46293650_46293801 | 0.06 |
CTD-2589M5.4 |
|
2373 |
0.27 |
| chr8_119820459_119820610 | 0.06 |
TNFRSF11B |
tumor necrosis factor receptor superfamily, member 11b |
143905 |
0.05 |
| chr3_73552092_73552243 | 0.06 |
PDZRN3 |
PDZ domain containing ring finger 3 |
28384 |
0.23 |
| chr1_84842738_84842889 | 0.06 |
DNASE2B |
deoxyribonuclease II beta |
21402 |
0.19 |
| chr16_59670319_59670470 | 0.06 |
ENSG00000200062 |
. |
96364 |
0.09 |
| chr9_116394736_116394887 | 0.06 |
RGS3 |
regulator of G-protein signaling 3 |
39045 |
0.17 |
| chr3_187989157_187989308 | 0.06 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
31580 |
0.22 |
| chr6_144007887_144008177 | 0.06 |
PHACTR2 |
phosphatase and actin regulator 2 |
8827 |
0.27 |
| chr1_51730316_51730746 | 0.06 |
RP11-296A18.6 |
|
1215 |
0.4 |
| chr12_52575126_52575277 | 0.06 |
KRT80 |
keratin 80 |
10583 |
0.13 |
| chr11_123460981_123461132 | 0.06 |
GRAMD1B |
GRAM domain containing 1B |
17461 |
0.21 |
| chr6_33580965_33581212 | 0.06 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
7054 |
0.14 |
| chr2_227624493_227624644 | 0.06 |
IRS1 |
insulin receptor substrate 1 |
39907 |
0.16 |
| chr3_49842732_49843503 | 0.06 |
ENSG00000263506 |
. |
561 |
0.55 |
| chr11_2333873_2334024 | 0.06 |
ENSG00000207308 |
. |
9000 |
0.12 |
| chr16_88703444_88703870 | 0.06 |
IL17C |
interleukin 17C |
1344 |
0.26 |
| chr3_123838707_123838915 | 0.06 |
ENSG00000266383 |
. |
12965 |
0.21 |
| chr3_149325355_149325506 | 0.06 |
WWTR1 |
WW domain containing transcription regulator 1 |
31391 |
0.15 |
| chr9_112942282_112942517 | 0.06 |
C9orf152 |
chromosome 9 open reading frame 152 |
28070 |
0.2 |
| chr20_10963665_10963956 | 0.06 |
RP11-103J8.1 |
|
3513 |
0.34 |
| chr5_139069430_139069742 | 0.05 |
CXXC5 |
CXXC finger protein 5 |
10318 |
0.2 |
| chr10_126745390_126745757 | 0.05 |
ENSG00000264572 |
. |
24134 |
0.19 |
| chr2_129063259_129063630 | 0.05 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
12707 |
0.24 |
| chr6_19698381_19698532 | 0.05 |
ENSG00000200957 |
. |
55696 |
0.16 |
| chr12_47025316_47025467 | 0.05 |
SLC38A4 |
solute carrier family 38, member 4 |
137696 |
0.05 |
| chr10_22723256_22723780 | 0.05 |
RP11-301N24.3 |
|
73417 |
0.1 |
| chr8_118826933_118827084 | 0.05 |
MED30 |
mediator complex subunit 30 |
293936 |
0.01 |
| chr2_133157691_133157842 | 0.05 |
ENSG00000252705 |
. |
6665 |
0.23 |
| chr11_74037423_74037574 | 0.05 |
P4HA3 |
prolyl 4-hydroxylase, alpha polypeptide III |
14796 |
0.16 |
| chr17_15224796_15224947 | 0.05 |
ENSG00000200437 |
. |
10209 |
0.17 |
| chr3_154688977_154689233 | 0.05 |
MME |
membrane metallo-endopeptidase |
52808 |
0.18 |
| chr4_120573726_120573877 | 0.05 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
23655 |
0.24 |
| chr3_99129524_99129675 | 0.05 |
ENSG00000266030 |
. |
43866 |
0.2 |
| chr8_99164226_99164377 | 0.05 |
POP1 |
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
34233 |
0.13 |
| chr11_70634178_70634329 | 0.05 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
38392 |
0.16 |
| chr2_69449091_69449242 | 0.05 |
ENSG00000199460 |
. |
39157 |
0.15 |
| chr6_122329509_122329660 | 0.05 |
ENSG00000199932 |
. |
203210 |
0.03 |
| chr10_125689568_125689719 | 0.05 |
RP11-285A18.2 |
|
1470 |
0.43 |
| chr13_111059030_111059181 | 0.05 |
ENSG00000238629 |
. |
7447 |
0.24 |
| chr7_90447196_90447347 | 0.05 |
CDK14 |
cyclin-dependent kinase 14 |
108095 |
0.07 |
| chrX_119003137_119003794 | 0.05 |
NDUFA1 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa |
1985 |
0.21 |
| chr8_123607926_123608077 | 0.05 |
ENSG00000238901 |
. |
75529 |
0.12 |
| chr14_51974464_51974948 | 0.05 |
ENSG00000252400 |
. |
14876 |
0.21 |
| chr3_55367534_55367685 | 0.05 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
147615 |
0.05 |
| chr21_44922828_44923116 | 0.05 |
SIK1 |
salt-inducible kinase 1 |
75964 |
0.1 |
| chr2_216764593_216764988 | 0.05 |
ENSG00000212055 |
. |
21148 |
0.25 |
| chr22_46402793_46403252 | 0.05 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
30013 |
0.1 |
| chr2_46612633_46612855 | 0.05 |
ENSG00000241791 |
. |
62906 |
0.11 |
| chr2_216613985_216614378 | 0.05 |
ENSG00000212055 |
. |
129461 |
0.05 |
| chr14_94587792_94587943 | 0.05 |
IFI27L2 |
interferon, alpha-inducible protein 27-like 2 |
8134 |
0.15 |
| chr15_52935123_52935274 | 0.05 |
RP11-519C12.1 |
|
6865 |
0.2 |
| chr9_118294239_118294390 | 0.05 |
DEC1 |
deleted in esophageal cancer 1 |
390217 |
0.01 |
| chr6_158506907_158507345 | 0.05 |
SYNJ2 |
synaptojanin 2 |
8414 |
0.21 |
| chr9_130615331_130616090 | 0.05 |
ENG |
endoglin |
1205 |
0.25 |
| chr6_140075327_140075478 | 0.05 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
379645 |
0.01 |
| chr18_11323846_11323997 | 0.05 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
175334 |
0.03 |
| chr11_95555093_95555244 | 0.05 |
CEP57 |
centrosomal protein 57kDa |
4167 |
0.24 |
| chr3_99964733_99964884 | 0.05 |
TBC1D23 |
TBC1 domain family, member 23 |
15036 |
0.17 |
| chr15_66938215_66938421 | 0.05 |
SMAD6 |
SMAD family member 6 |
56248 |
0.1 |
| chr1_183041587_183041738 | 0.05 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
24017 |
0.19 |
| chr10_63553300_63553451 | 0.05 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
107684 |
0.07 |
| chr10_15336257_15336446 | 0.05 |
RP11-25G10.2 |
|
56860 |
0.13 |
| chr6_151151372_151151523 | 0.05 |
MTHFD1L |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
35238 |
0.2 |
| chr17_13496614_13496765 | 0.05 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
8555 |
0.26 |
| chr7_134380957_134381211 | 0.05 |
BPGM |
2,3-bisphosphoglycerate mutase |
35911 |
0.18 |
| chr6_36216557_36216942 | 0.05 |
PNPLA1 |
patatin-like phospholipase domain containing 1 |
5769 |
0.2 |
| chr17_75251415_75251576 | 0.05 |
SEPT9 |
septin 9 |
25156 |
0.19 |
| chr11_69325521_69325672 | 0.05 |
CCND1 |
cyclin D1 |
130259 |
0.05 |
| chr6_85787065_85787216 | 0.05 |
TBX18 |
T-box 18 |
312903 |
0.01 |
| chr11_70228081_70228232 | 0.05 |
PPFIA1 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
3972 |
0.15 |
| chr11_130761204_130761355 | 0.05 |
SNX19 |
sorting nexin 19 |
2490 |
0.42 |
| chr3_66456383_66456534 | 0.05 |
ENSG00000241587 |
. |
82179 |
0.1 |
| chr12_2394240_2394442 | 0.05 |
CACNA1C-IT3 |
CACNA1C intronic transcript 3 (non-protein coding) |
15399 |
0.25 |
| chr9_98260446_98260655 | 0.05 |
RP11-435O5.6 |
|
3344 |
0.21 |
| chr1_21066214_21066365 | 0.05 |
RP5-930J4.4 |
|
3191 |
0.17 |
| chr20_45754813_45755050 | 0.05 |
ENSG00000264901 |
. |
40678 |
0.18 |
| chr14_53301825_53302222 | 0.05 |
FERMT2 |
fermitin family member 2 |
29216 |
0.15 |
| chr9_74545305_74545456 | 0.05 |
C9orf85 |
chromosome 9 open reading frame 85 |
18803 |
0.18 |
| chr2_40327696_40327847 | 0.05 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
4278 |
0.36 |
| chr5_171845939_171846090 | 0.05 |
ENSG00000216127 |
. |
20744 |
0.2 |
| chr16_86696225_86696376 | 0.05 |
FOXL1 |
forkhead box L1 |
84185 |
0.08 |
| chr1_235097341_235097798 | 0.05 |
ENSG00000239690 |
. |
57636 |
0.14 |
| chr1_113067186_113067337 | 0.05 |
RP4-671G15.2 |
|
6198 |
0.2 |
| chr7_94040475_94040626 | 0.05 |
COL1A2 |
collagen, type I, alpha 2 |
16677 |
0.25 |
| chr3_29724640_29724791 | 0.05 |
RBMS3-AS2 |
RBMS3 antisense RNA 2 |
40415 |
0.22 |
| chr7_72421339_72421490 | 0.05 |
NSUN5P2 |
NOP2/Sun domain family, member 5 pseudogene 2 |
3842 |
0.13 |
| chr5_39416035_39416186 | 0.05 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
8860 |
0.28 |
| chr10_81896535_81896686 | 0.05 |
PLAC9 |
placenta-specific 9 |
4133 |
0.22 |
| chr10_97033802_97033953 | 0.05 |
PDLIM1 |
PDZ and LIM domain 1 |
16904 |
0.21 |
| chr6_2378075_2378226 | 0.05 |
ENSG00000266252 |
. |
31719 |
0.24 |
| chr22_33104640_33104791 | 0.05 |
ENSG00000251890 |
. |
73531 |
0.1 |
| chr19_19458792_19458943 | 0.05 |
MAU2 |
MAU2 sister chromatid cohesion factor |
10852 |
0.13 |
| chr6_38315124_38315275 | 0.05 |
ENSG00000238716 |
. |
11806 |
0.28 |
| chr8_70555415_70555566 | 0.05 |
ENSG00000222889 |
. |
56969 |
0.13 |
| chr2_9450145_9450296 | 0.05 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
103326 |
0.06 |
| chr2_134599157_134599414 | 0.05 |
ENSG00000200708 |
. |
245623 |
0.02 |
| chr17_19233901_19234052 | 0.05 |
RP11-135L13.4 |
|
5464 |
0.12 |
| chr1_206687145_206687296 | 0.05 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
6341 |
0.15 |
| chr4_189862800_189862951 | 0.05 |
ENSG00000252275 |
. |
224996 |
0.02 |
| chr3_106329448_106329599 | 0.05 |
ENSG00000200361 |
. |
78002 |
0.11 |
| chr17_59310548_59310699 | 0.05 |
RP11-136H19.1 |
|
97298 |
0.07 |
| chr8_6344138_6344289 | 0.05 |
ANGPT2 |
angiopoietin 2 |
76352 |
0.1 |
| chr22_31039864_31040015 | 0.05 |
SLC35E4 |
solute carrier family 35, member E4 |
7429 |
0.13 |
| chr11_10139098_10139249 | 0.05 |
RP11-748C4.1 |
|
31566 |
0.21 |
| chr2_33363115_33363266 | 0.05 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
3466 |
0.36 |
| chr16_73156371_73156706 | 0.05 |
C16orf47 |
chromosome 16 open reading frame 47 |
21808 |
0.23 |
| chr6_52392885_52393036 | 0.05 |
TRAM2 |
translocation associated membrane protein 2 |
48753 |
0.15 |
| chr2_190030887_190031038 | 0.05 |
COL5A2 |
collagen, type V, alpha 2 |
13643 |
0.22 |
| chr7_98438304_98438455 | 0.05 |
TMEM130 |
transmembrane protein 130 |
26190 |
0.14 |
| chr8_24206460_24207135 | 0.05 |
ADAM28 |
ADAM metallopeptidase domain 28 |
7632 |
0.22 |
| chr10_80838963_80839341 | 0.05 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
10360 |
0.24 |
| chr1_223083321_223083472 | 0.05 |
ENSG00000239054 |
. |
1893 |
0.4 |
| chr9_104277002_104277153 | 0.05 |
TMEM246 |
transmembrane protein 246 |
18653 |
0.14 |
| chr1_6803545_6803777 | 0.05 |
ENSG00000239166 |
. |
9118 |
0.17 |
| chr10_31531410_31531796 | 0.05 |
ENSG00000252479 |
. |
16925 |
0.24 |
| chr6_64516560_64517136 | 0.05 |
RP11-59D5__B.2 |
|
119 |
0.98 |
| chr16_49673318_49673469 | 0.05 |
ZNF423 |
zinc finger protein 423 |
1370 |
0.54 |
| chr1_221437708_221437859 | 0.05 |
HLX |
H2.0-like homeobox |
383199 |
0.01 |
| chr11_122068067_122068218 | 0.05 |
ENSG00000207994 |
. |
45126 |
0.13 |
| chr11_113323627_113323778 | 0.05 |
ENSG00000265140 |
. |
2892 |
0.24 |
| chr19_38490719_38490870 | 0.05 |
CTC-244M17.1 |
|
36301 |
0.14 |
| chr1_210536298_210536449 | 0.05 |
HHAT |
hedgehog acyltransferase |
177 |
0.96 |
| chr20_19795494_19795742 | 0.05 |
AL121761.2 |
Uncharacterized protein |
56939 |
0.12 |
| chr2_106010411_106010562 | 0.05 |
FHL2 |
four and a half LIM domains 2 |
2756 |
0.27 |
| chr11_77654778_77654929 | 0.05 |
INTS4 |
integrator complex subunit 4 |
32653 |
0.12 |
| chr11_131729951_131730102 | 0.05 |
AP004372.1 |
|
36976 |
0.19 |
| chr1_159009390_159009541 | 0.05 |
ENSG00000265589 |
. |
19574 |
0.16 |
| chr10_80713188_80713394 | 0.05 |
ZMIZ1-AS1 |
ZMIZ1 antisense RNA 1 |
8860 |
0.31 |
| chr1_856648_857113 | 0.05 |
SAMD11 |
sterile alpha motif domain containing 11 |
3380 |
0.14 |
| chr1_53914957_53915108 | 0.05 |
DMRTB1 |
DMRT-like family B with proline-rich C-terminal, 1 |
10040 |
0.24 |
| chr11_61772142_61772440 | 0.05 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
36838 |
0.11 |
| chr15_63142711_63142998 | 0.05 |
RP11-1069G10.1 |
|
6074 |
0.22 |
| chr2_28884288_28884439 | 0.05 |
AC074011.2 |
|
3043 |
0.24 |
| chr2_10119571_10119722 | 0.05 |
GRHL1 |
grainyhead-like 1 (Drosophila) |
15788 |
0.16 |
| chr6_126135548_126136266 | 0.05 |
NCOA7 |
nuclear receptor coactivator 7 |
457 |
0.81 |
| chr18_48596659_48596948 | 0.05 |
SMAD4 |
SMAD family member 4 |
3320 |
0.29 |
| chr9_111815301_111815452 | 0.05 |
ENSG00000207698 |
. |
6798 |
0.16 |
| chr14_55603634_55603785 | 0.05 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
7731 |
0.23 |
| chr16_64447214_64447365 | 0.05 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
646292 |
0.0 |
| chr9_16031542_16031875 | 0.05 |
C9orf92 |
chromosome 9 open reading frame 92 |
184189 |
0.03 |
| chr6_38314206_38314357 | 0.05 |
ENSG00000238716 |
. |
12724 |
0.28 |
| chr7_75043431_75043582 | 0.05 |
NSUN5P1 |
NOP2/Sun domain family, member 5 pseudogene 1 |
1676 |
0.23 |
| chr19_42781870_42782547 | 0.05 |
CIC |
capicua transcriptional repressor |
5964 |
0.09 |
| chr6_113468381_113468532 | 0.05 |
ENSG00000201386 |
. |
175761 |
0.03 |
| chr14_71710075_71710226 | 0.05 |
AC004817.1 |
HCG2028611; Uncharacterized protein |
26688 |
0.21 |
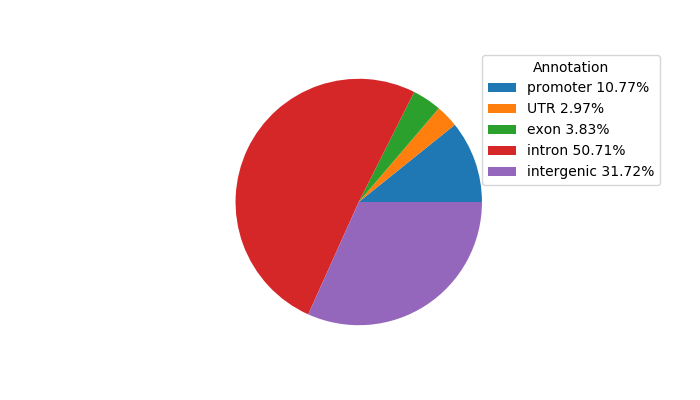
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |