Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
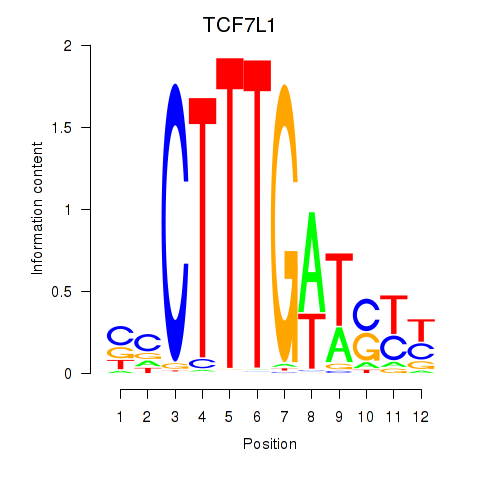
Results for TCF7L1
Z-value: 0.93
Transcription factors associated with TCF7L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L1
|
ENSG00000152284.4 | transcription factor 7 like 1 |
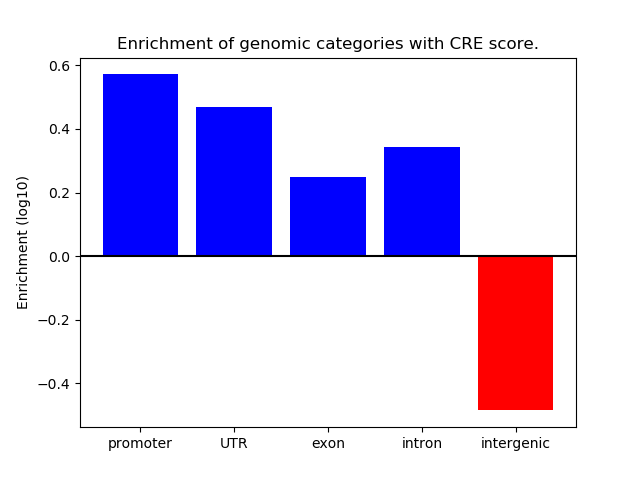
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_85284511_85284681 | TCF7L1 | 75937 | 0.084333 | 0.77 | 1.6e-02 | Click! |
| chr2_85282591_85282742 | TCF7L1 | 77867 | 0.081553 | -0.60 | 8.9e-02 | Click! |
| chr2_85284961_85285173 | TCF7L1 | 75466 | 0.085025 | 0.56 | 1.2e-01 | Click! |
| chr2_85359450_85359601 | TCF7L1 | 1008 | 0.580287 | 0.55 | 1.2e-01 | Click! |
| chr2_85280075_85280226 | TCF7L1 | 80383 | 0.078064 | 0.46 | 2.1e-01 | Click! |
Activity of the TCF7L1 motif across conditions
Conditions sorted by the z-value of the TCF7L1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
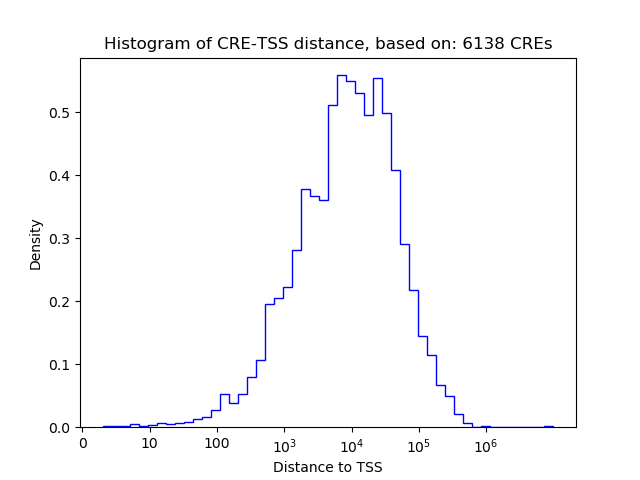
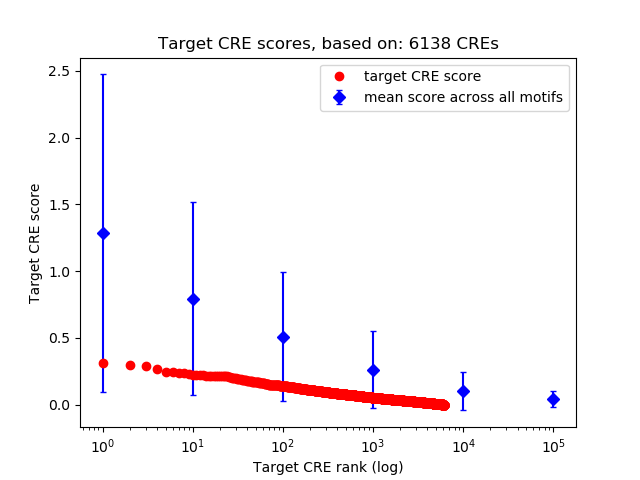
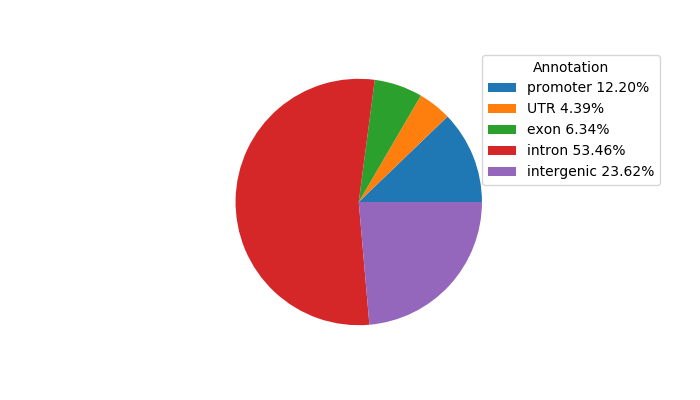
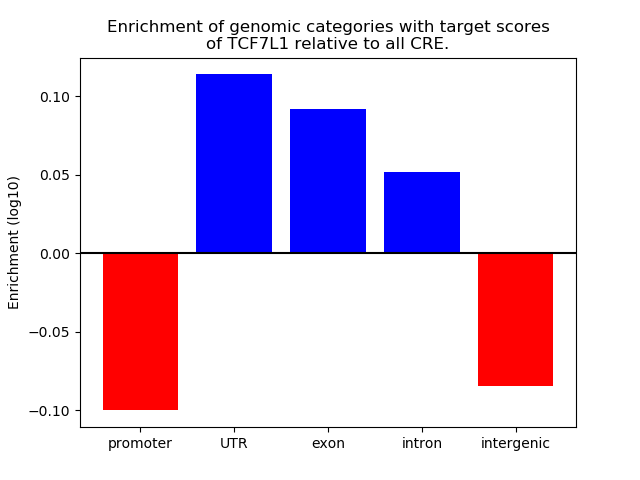
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_49946029_49946413 | 0.31 |
CAB39L |
calcium binding protein 39-like |
5046 |
0.26 |
| chr17_38182542_38182693 | 0.30 |
ENSG00000238793 |
. |
1281 |
0.3 |
| chr14_68648383_68648543 | 0.29 |
ENSG00000244677 |
. |
31838 |
0.18 |
| chr16_50651385_50651565 | 0.27 |
RP11-401P9.6 |
|
3880 |
0.2 |
| chr8_128840086_128840237 | 0.25 |
ENSG00000249859 |
. |
31953 |
0.2 |
| chr2_109580892_109581113 | 0.24 |
EDAR |
ectodysplasin A receptor |
24723 |
0.24 |
| chr5_156319485_156319636 | 0.24 |
TIMD4 |
T-cell immunoglobulin and mucin domain containing 4 |
43127 |
0.17 |
| chr11_130259035_130259186 | 0.24 |
ADAMTS8 |
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
39778 |
0.16 |
| chr5_76625160_76625430 | 0.23 |
PDE8B |
phosphodiesterase 8B |
3777 |
0.34 |
| chr11_57449245_57449396 | 0.22 |
ZDHHC5 |
zinc finger, DHHC-type containing 5 |
692 |
0.53 |
| chr3_49025329_49025480 | 0.22 |
P4HTM |
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
1915 |
0.14 |
| chr12_65094836_65094987 | 0.22 |
AC025262.1 |
Mesenchymal stem cell protein DSC96; Uncharacterized protein |
4582 |
0.17 |
| chr4_4227781_4227932 | 0.22 |
OTOP1 |
otopetrin 1 |
760 |
0.67 |
| chr10_73862702_73862853 | 0.22 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
13987 |
0.21 |
| chr4_981402_981614 | 0.22 |
IDUA |
iduronidase, alpha-L- |
633 |
0.46 |
| chr1_207073979_207074130 | 0.22 |
IL24 |
interleukin 24 |
2876 |
0.21 |
| chr19_33662270_33662421 | 0.22 |
ENSG00000264355 |
. |
5633 |
0.14 |
| chr9_98620223_98620374 | 0.22 |
ENSG00000252847 |
. |
1888 |
0.36 |
| chr7_100814110_100814261 | 0.22 |
VGF |
VGF nerve growth factor inducible |
5311 |
0.1 |
| chr13_22681618_22681904 | 0.22 |
ENSG00000244197 |
. |
317085 |
0.01 |
| chr14_65237977_65238128 | 0.21 |
SPTB |
spectrin, beta, erythrocytic |
11216 |
0.2 |
| chr18_67070231_67070382 | 0.21 |
DOK6 |
docking protein 6 |
2015 |
0.5 |
| chr17_1776866_1777017 | 0.21 |
RPA1 |
replication protein A1, 70kDa |
5388 |
0.17 |
| chr2_235056234_235056385 | 0.21 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
96878 |
0.08 |
| chr15_87160145_87160540 | 0.21 |
RP11-182L7.1 |
|
14025 |
0.31 |
| chr16_1018736_1018887 | 0.21 |
LMF1 |
lipase maturation factor 1 |
2055 |
0.21 |
| chr12_124416244_124416395 | 0.20 |
DNAH10OS |
dynein, axonemal, heavy chain 10 opposite strand |
3212 |
0.11 |
| chr16_3064160_3064311 | 0.20 |
CLDN9 |
claudin 9 |
1778 |
0.12 |
| chr7_27278726_27278948 | 0.20 |
EVX1 |
even-skipped homeobox 1 |
3327 |
0.13 |
| chr13_30532983_30533134 | 0.20 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
32270 |
0.23 |
| chr11_62285356_62285507 | 0.20 |
AHNAK |
AHNAK nucleoprotein |
18148 |
0.09 |
| chr7_537977_538128 | 0.19 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
20093 |
0.2 |
| chr8_19250899_19251050 | 0.19 |
SH2D4A |
SH2 domain containing 4A |
73873 |
0.13 |
| chr2_99527888_99528039 | 0.19 |
KIAA1211L |
KIAA1211-like |
2979 |
0.33 |
| chr1_111212908_111213059 | 0.19 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
4672 |
0.21 |
| chr5_80400951_80401102 | 0.19 |
CTD-2193P3.2 |
|
9645 |
0.28 |
| chr1_46808965_46809116 | 0.19 |
NSUN4 |
NOP2/Sun domain family, member 4 |
2164 |
0.27 |
| chr7_141211304_141211601 | 0.18 |
AGK |
acylglycerol kinase |
39537 |
0.14 |
| chr16_69200425_69200916 | 0.18 |
ENSG00000207083 |
. |
9010 |
0.12 |
| chr11_13317651_13317839 | 0.18 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
13788 |
0.26 |
| chr2_25498362_25498513 | 0.18 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
23257 |
0.19 |
| chr1_93042440_93042924 | 0.18 |
GFI1 |
growth factor independent 1 transcription repressor |
90249 |
0.08 |
| chr1_168482288_168482558 | 0.18 |
XCL2 |
chemokine (C motif) ligand 2 |
30812 |
0.2 |
| chr7_157619868_157620019 | 0.18 |
AC011899.9 |
|
27278 |
0.2 |
| chr21_34557928_34558079 | 0.17 |
C21orf54 |
chromosome 21 open reading frame 54 |
15462 |
0.15 |
| chr10_105564418_105564569 | 0.17 |
SH3PXD2A |
SH3 and PX domains 2A |
3438 |
0.29 |
| chr20_415273_415424 | 0.17 |
ENSG00000206797 |
. |
15959 |
0.14 |
| chr6_111908214_111908423 | 0.17 |
TRAF3IP2-AS1 |
TRAF3IP2 antisense RNA 1 |
12496 |
0.17 |
| chr2_237283808_237283959 | 0.17 |
RP11-785G17.1 |
|
7680 |
0.22 |
| chr8_66770156_66770307 | 0.17 |
PDE7A |
phosphodiesterase 7A |
16044 |
0.27 |
| chr6_143996619_143996770 | 0.17 |
PHACTR2 |
phosphatase and actin regulator 2 |
2408 |
0.39 |
| chr19_42744288_42744439 | 0.17 |
GSK3A |
glycogen synthase kinase 3 alpha |
1682 |
0.17 |
| chr9_123665604_123665920 | 0.17 |
TRAF1 |
TNF receptor-associated factor 1 |
11088 |
0.19 |
| chr7_43651721_43652066 | 0.17 |
STK17A |
serine/threonine kinase 17a |
29229 |
0.15 |
| chr5_172464440_172464591 | 0.17 |
ENSG00000207210 |
. |
15043 |
0.14 |
| chr9_20640905_20641056 | 0.16 |
FOCAD |
focadhesin |
17328 |
0.18 |
| chr2_231279331_231279482 | 0.16 |
SP100 |
SP100 nuclear antigen |
1251 |
0.55 |
| chr1_12674943_12675094 | 0.16 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
2719 |
0.25 |
| chr9_130735456_130735638 | 0.16 |
FAM102A |
family with sequence similarity 102, member A |
7245 |
0.11 |
| chr3_13033156_13033307 | 0.16 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
4695 |
0.3 |
| chr7_143002225_143002376 | 0.16 |
ENSG00000240322 |
. |
6691 |
0.1 |
| chr17_8799606_8799757 | 0.16 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
14495 |
0.2 |
| chr6_33597995_33598146 | 0.16 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
8909 |
0.14 |
| chr2_106711761_106712080 | 0.16 |
C2orf40 |
chromosome 2 open reading frame 40 |
29664 |
0.2 |
| chr19_16708392_16708543 | 0.16 |
CTD-3222D19.5 |
|
9671 |
0.09 |
| chr21_34666749_34666900 | 0.15 |
IL10RB |
interleukin 10 receptor, beta |
28161 |
0.12 |
| chr3_108547468_108547619 | 0.15 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
5924 |
0.28 |
| chr9_138999698_138999849 | 0.15 |
C9orf69 |
chromosome 9 open reading frame 69 |
10347 |
0.19 |
| chr3_107694284_107694711 | 0.15 |
CD47 |
CD47 molecule |
82711 |
0.11 |
| chr5_133382798_133382949 | 0.15 |
VDAC1 |
voltage-dependent anion channel 1 |
42049 |
0.15 |
| chr6_157119654_157119805 | 0.15 |
RP11-230C9.3 |
|
18140 |
0.19 |
| chr13_96743482_96743694 | 0.15 |
HS6ST3 |
heparan sulfate 6-O-sulfotransferase 3 |
495 |
0.85 |
| chr9_96327899_96328050 | 0.15 |
ENSG00000263875 |
. |
108 |
0.97 |
| chr8_121783196_121783347 | 0.15 |
RP11-713M15.1 |
|
9778 |
0.23 |
| chr17_39496038_39496239 | 0.15 |
KRT33A |
keratin 33A |
10926 |
0.08 |
| chr2_179910796_179910947 | 0.15 |
CCDC141 |
coiled-coil domain containing 141 |
3915 |
0.27 |
| chr16_87876134_87876285 | 0.15 |
RP4-536B24.2 |
|
6071 |
0.19 |
| chr21_45359193_45359344 | 0.15 |
AGPAT3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
7543 |
0.18 |
| chr3_9829316_9829467 | 0.15 |
ARPC4 |
actin related protein 2/3 complex, subunit 4, 20kDa |
4788 |
0.1 |
| chr7_158571577_158571883 | 0.15 |
ESYT2 |
extended synaptotagmin-like protein 2 |
34822 |
0.19 |
| chr22_31502188_31502378 | 0.15 |
SELM |
Selenoprotein M |
1271 |
0.3 |
| chr5_86709837_86709988 | 0.15 |
CCNH |
cyclin H |
1076 |
0.53 |
| chr8_8197619_8197770 | 0.15 |
SGK223 |
Tyrosine-protein kinase SgK223 |
41563 |
0.16 |
| chr5_141821544_141821695 | 0.15 |
AC005592.2 |
|
37836 |
0.19 |
| chr14_22959333_22959484 | 0.15 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
3237 |
0.13 |
| chr10_75953508_75953715 | 0.15 |
ADK |
adenosine kinase |
17090 |
0.22 |
| chr15_70540546_70541057 | 0.15 |
ENSG00000200216 |
. |
55226 |
0.16 |
| chr8_134494949_134495197 | 0.15 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
6226 |
0.32 |
| chr2_120595579_120595730 | 0.15 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
77854 |
0.1 |
| chr10_8406671_8406822 | 0.15 |
ENSG00000212505 |
. |
292048 |
0.01 |
| chr19_42426367_42426518 | 0.14 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
804 |
0.52 |
| chr5_176816529_176816680 | 0.14 |
SLC34A1 |
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
5159 |
0.1 |
| chr6_42291647_42291798 | 0.14 |
ENSG00000221252 |
. |
88412 |
0.07 |
| chr1_8467782_8467980 | 0.14 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
15845 |
0.18 |
| chr3_51312232_51312383 | 0.14 |
MANF |
mesencephalic astrocyte-derived neurotrophic factor |
110171 |
0.06 |
| chr10_3494380_3494531 | 0.14 |
PITRM1 |
pitrilysin metallopeptidase 1 |
279452 |
0.01 |
| chr13_114931895_114932090 | 0.14 |
RASA3 |
RAS p21 protein activator 3 |
33906 |
0.16 |
| chr4_148767939_148768090 | 0.14 |
ARHGAP10 |
Rho GTPase activating protein 10 |
35004 |
0.16 |
| chr3_16414400_16414551 | 0.14 |
RP11-415F23.4 |
|
27842 |
0.17 |
| chr15_63885452_63885603 | 0.14 |
USP3-AS1 |
USP3 antisense RNA 1 |
1376 |
0.42 |
| chr10_105189029_105189180 | 0.14 |
RP11-225H22.7 |
|
20849 |
0.09 |
| chr12_69285926_69286077 | 0.14 |
ENSG00000252770 |
. |
19700 |
0.15 |
| chr11_125955059_125955210 | 0.14 |
CDON |
cell adhesion associated, oncogene regulated |
21904 |
0.18 |
| chr13_51170641_51170830 | 0.14 |
DLEU7-AS1 |
DLEU7 antisense RNA 1 |
211257 |
0.02 |
| chr10_112216760_112216911 | 0.14 |
ENSG00000264544 |
. |
18517 |
0.17 |
| chr2_235337973_235338124 | 0.14 |
ARL4C |
ADP-ribosylation factor-like 4C |
67196 |
0.14 |
| chr7_129417298_129417546 | 0.14 |
ENSG00000207691 |
. |
2568 |
0.21 |
| chr6_134595465_134595616 | 0.14 |
ENSG00000201309 |
. |
8690 |
0.19 |
| chr22_47184851_47185002 | 0.14 |
TBC1D22A |
TBC1 domain family, member 22A |
15102 |
0.19 |
| chr17_7163807_7164082 | 0.14 |
CLDN7 |
claudin 7 |
520 |
0.48 |
| chr7_45958016_45958250 | 0.14 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
1108 |
0.52 |
| chr1_36003475_36003698 | 0.14 |
KIAA0319L |
KIAA0319-like |
17002 |
0.14 |
| chr5_126406425_126406576 | 0.14 |
C5orf63 |
chromosome 5 open reading frame 63 |
815 |
0.72 |
| chr22_30692525_30692676 | 0.14 |
RP1-130H16.18 |
Uncharacterized protein |
2871 |
0.14 |
| chr13_26480038_26480189 | 0.13 |
AL138815.2 |
Uncharacterized protein |
27434 |
0.16 |
| chr10_12222611_12222762 | 0.13 |
NUDT5 |
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
15135 |
0.12 |
| chr4_48355584_48355735 | 0.13 |
SLAIN2 |
SLAIN motif family, member 2 |
12320 |
0.25 |
| chr2_175615864_175616015 | 0.13 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
13201 |
0.24 |
| chr5_14415059_14415274 | 0.13 |
TRIO |
trio Rho guanine nucleotide exchange factor |
73391 |
0.12 |
| chr12_103356683_103356834 | 0.13 |
PAH |
phenylalanine hydroxylase |
4614 |
0.22 |
| chr20_3783093_3783244 | 0.13 |
CDC25B |
cell division cycle 25B |
6090 |
0.12 |
| chr10_21785028_21785179 | 0.13 |
ENSG00000222071 |
. |
467 |
0.64 |
| chr6_157194356_157194507 | 0.13 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
28076 |
0.23 |
| chr3_71532445_71532638 | 0.13 |
ENSG00000221264 |
. |
58699 |
0.13 |
| chr1_167453393_167453610 | 0.13 |
RP11-104L21.2 |
|
25603 |
0.17 |
| chr3_128835970_128836121 | 0.13 |
RAB43 |
RAB43, member RAS oncogene family |
4579 |
0.14 |
| chr14_100579946_100580097 | 0.13 |
ENSG00000199082 |
. |
4029 |
0.15 |
| chr1_116929404_116929555 | 0.13 |
AL136376.1 |
Uncharacterized protein |
2761 |
0.21 |
| chr18_51804642_51804943 | 0.13 |
POLI |
polymerase (DNA directed) iota |
4484 |
0.25 |
| chr1_214156856_214157007 | 0.13 |
PROX1 |
prospero homeobox 1 |
407 |
0.85 |
| chr11_61133550_61133701 | 0.13 |
TMEM138 |
transmembrane protein 138 |
450 |
0.71 |
| chr11_105886093_105886244 | 0.13 |
MSANTD4 |
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
6786 |
0.2 |
| chrX_117674478_117674629 | 0.13 |
ENSG00000206862 |
. |
28925 |
0.2 |
| chr17_1081161_1081317 | 0.13 |
ABR |
active BCR-related |
1839 |
0.34 |
| chr17_2407698_2407849 | 0.13 |
METTL16 |
methyltransferase like 16 |
7407 |
0.18 |
| chr1_186258649_186258839 | 0.13 |
PRG4 |
proteoglycan 4 |
6661 |
0.2 |
| chr19_47947885_47948253 | 0.13 |
MEIS3 |
Meis homeobox 3 |
25289 |
0.11 |
| chr9_132393459_132393610 | 0.13 |
NTMT1 |
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
4811 |
0.13 |
| chr3_108553144_108553369 | 0.13 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
11637 |
0.26 |
| chr1_12530168_12530319 | 0.13 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
5248 |
0.24 |
| chr15_31642037_31642188 | 0.13 |
KLF13 |
Kruppel-like factor 13 |
9976 |
0.3 |
| chr2_240085793_240086170 | 0.13 |
AC017028.1 |
Uncharacterized protein |
1928 |
0.27 |
| chr5_156473476_156473627 | 0.13 |
HAVCR1 |
hepatitis A virus cellular receptor 1 |
11856 |
0.19 |
| chr11_35353117_35353423 | 0.13 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
30195 |
0.17 |
| chr16_58154872_58155238 | 0.12 |
CTB-134F13.1 |
|
4266 |
0.14 |
| chr15_31495377_31495528 | 0.12 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
41976 |
0.18 |
| chr1_94081045_94081360 | 0.12 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
1548 |
0.43 |
| chrX_40013584_40013785 | 0.12 |
BCOR |
BCL6 corepressor |
7802 |
0.32 |
| chr10_95460096_95460247 | 0.12 |
FRA10AC1 |
fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 |
2154 |
0.35 |
| chr11_128381097_128381643 | 0.12 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
6081 |
0.25 |
| chr5_16673046_16673197 | 0.12 |
ENSG00000201370 |
. |
49917 |
0.12 |
| chr7_48084885_48085036 | 0.12 |
C7orf57 |
chromosome 7 open reading frame 57 |
9843 |
0.19 |
| chr5_109280950_109281101 | 0.12 |
AC011366.3 |
Uncharacterized protein |
62142 |
0.16 |
| chr14_89890480_89890631 | 0.12 |
FOXN3-AS1 |
FOXN3 antisense RNA 1 |
6857 |
0.18 |
| chr7_156795968_156796187 | 0.12 |
MNX1-AS2 |
MNX1 antisense RNA 2 |
2924 |
0.22 |
| chr12_56685183_56685363 | 0.12 |
CS |
citrate synthase |
8485 |
0.07 |
| chr6_91123599_91123783 | 0.12 |
ENSG00000252676 |
. |
32800 |
0.22 |
| chr12_3563578_3563841 | 0.12 |
RP11-476M19.3 |
|
9043 |
0.26 |
| chr11_64088032_64088183 | 0.12 |
PRDX5 |
peroxiredoxin 5 |
2453 |
0.1 |
| chr5_109193615_109193766 | 0.12 |
AC011366.3 |
Uncharacterized protein |
25193 |
0.26 |
| chr2_128391312_128391493 | 0.12 |
RP11-286H15.1 |
|
6979 |
0.14 |
| chr19_42396256_42396453 | 0.12 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
2011 |
0.21 |
| chr9_135036094_135036322 | 0.12 |
NTNG2 |
netrin G2 |
1126 |
0.59 |
| chr1_166815002_166815153 | 0.12 |
POGK |
pogo transposable element with KRAB domain |
5751 |
0.25 |
| chr11_59378018_59378169 | 0.12 |
OSBP |
oxysterol binding protein |
5524 |
0.13 |
| chr21_43744243_43744394 | 0.12 |
TFF3 |
trefoil factor 3 (intestinal) |
8557 |
0.14 |
| chr6_112092329_112092498 | 0.12 |
FYN |
FYN oncogene related to SRC, FGR, YES |
9870 |
0.28 |
| chr6_159439729_159439953 | 0.12 |
RP1-111C20.4 |
|
7137 |
0.19 |
| chr4_109062726_109063018 | 0.12 |
LEF1 |
lymphoid enhancer-binding factor 1 |
24585 |
0.2 |
| chr16_89397725_89397876 | 0.12 |
ANKRD11 |
ankyrin repeat domain 11 |
2406 |
0.21 |
| chr7_150676591_150676887 | 0.12 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
1336 |
0.32 |
| chr10_121160302_121160453 | 0.12 |
GRK5 |
G protein-coupled receptor kinase 5 |
22428 |
0.18 |
| chr2_159600251_159600402 | 0.12 |
AC005042.4 |
|
8812 |
0.25 |
| chr12_54440337_54440488 | 0.12 |
HOXC4 |
homeobox C4 |
7249 |
0.08 |
| chr12_92522877_92523028 | 0.12 |
C12orf79 |
chromosome 12 open reading frame 79 |
7845 |
0.19 |
| chr19_10520829_10520980 | 0.12 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
6545 |
0.11 |
| chr10_11324434_11324585 | 0.12 |
CELF2-AS1 |
CELF2 antisense RNA 1 |
37338 |
0.2 |
| chr6_170747344_170747495 | 0.12 |
RP1-140C12.2 |
|
24192 |
0.21 |
| chr1_186274017_186274168 | 0.12 |
ENSG00000202025 |
. |
6968 |
0.19 |
| chr1_11415481_11415632 | 0.11 |
UBIAD1 |
UbiA prenyltransferase domain containing 1 |
81565 |
0.08 |
| chr19_16486076_16486459 | 0.11 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
13503 |
0.14 |
| chr5_96447388_96447539 | 0.11 |
CTD-2215E18.1 |
Uncharacterized protein |
22684 |
0.17 |
| chr1_153941744_153941895 | 0.11 |
CREB3L4 |
cAMP responsive element binding protein 3-like 4 |
1033 |
0.26 |
| chr9_134153649_134153927 | 0.11 |
FAM78A |
family with sequence similarity 78, member A |
1854 |
0.33 |
| chr2_240169500_240169676 | 0.11 |
HDAC4 |
histone deacetylase 4 |
56836 |
0.1 |
| chr1_226848006_226848395 | 0.11 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
829 |
0.65 |
| chr4_4297415_4297566 | 0.11 |
ZBTB49 |
zinc finger and BTB domain containing 49 |
5494 |
0.19 |
| chr7_128043233_128043384 | 0.11 |
IMPDH1 |
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
2688 |
0.21 |
| chr14_98689187_98689819 | 0.11 |
ENSG00000222066 |
. |
108584 |
0.07 |
| chr2_106722867_106723018 | 0.11 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
32300 |
0.18 |
| chr16_89461717_89461868 | 0.11 |
RP1-168P16.2 |
|
17180 |
0.13 |
| chr2_24836758_24836909 | 0.11 |
NCOA1 |
nuclear receptor coactivator 1 |
29423 |
0.18 |
| chr1_117937222_117937373 | 0.11 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
25922 |
0.25 |
| chr17_8194100_8194251 | 0.11 |
RANGRF |
RAN guanine nucleotide release factor |
2179 |
0.15 |
| chrY_23267845_23267996 | 0.11 |
CYorf17 |
chromosome Y open reading frame 17 |
280326 |
0.01 |
| chr1_32294818_32294969 | 0.11 |
SPOCD1 |
SPOC domain containing 1 |
13241 |
0.16 |
| chr8_134300829_134300980 | 0.11 |
NDRG1 |
N-myc downstream regulated 1 |
7875 |
0.26 |
| chr12_115121082_115121268 | 0.11 |
TBX3 |
T-box 3 |
220 |
0.95 |
| chr2_84505605_84505756 | 0.11 |
SUCLG1 |
succinate-CoA ligase, alpha subunit |
180924 |
0.03 |
| chr3_149375939_149376119 | 0.11 |
WWTR1-AS1 |
WWTR1 antisense RNA 1 |
32 |
0.63 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0072193 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.0 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.0 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0034595 | phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |