Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
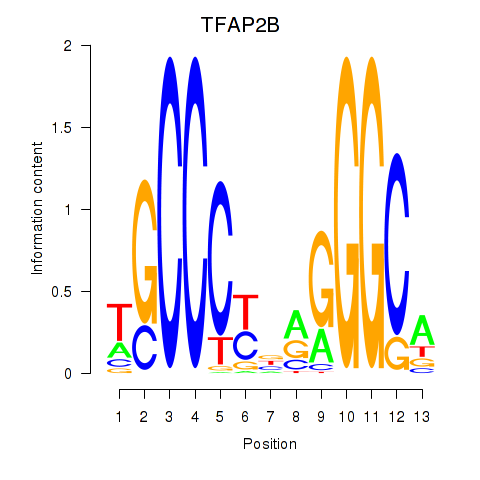
Results for TFAP2B
Z-value: 1.60
Transcription factors associated with TFAP2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2B
|
ENSG00000008196.8 | transcription factor AP-2 beta |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_50783493_50783702 | TFAP2B | 2839 | 0.410011 | -0.82 | 6.2e-03 | Click! |
| chr6_50903539_50903690 | TFAP2B | 117175 | 0.068877 | -0.79 | 1.2e-02 | Click! |
| chr6_50929546_50929733 | TFAP2B | 143200 | 0.049753 | -0.76 | 1.7e-02 | Click! |
| chr6_50929115_50929282 | TFAP2B | 142759 | 0.050012 | -0.69 | 3.9e-02 | Click! |
| chr6_50789163_50789314 | TFAP2B | 2799 | 0.414549 | 0.69 | 4.1e-02 | Click! |
Activity of the TFAP2B motif across conditions
Conditions sorted by the z-value of the TFAP2B motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
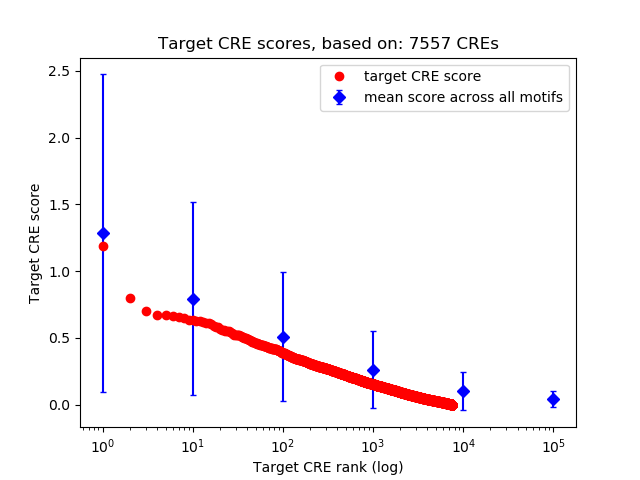
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_228336809_228337095 | 1.19 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
601 |
0.58 |
| chr9_133302644_133302795 | 0.80 |
HMCN2 |
hemicentin 2 |
2950 |
0.21 |
| chr10_112168315_112168466 | 0.70 |
ENSG00000221359 |
. |
25609 |
0.19 |
| chr7_4771612_4771763 | 0.67 |
FOXK1 |
forkhead box K1 |
8803 |
0.19 |
| chr19_4713192_4713546 | 0.67 |
DPP9 |
dipeptidyl-peptidase 9 |
949 |
0.43 |
| chr15_74679279_74679430 | 0.66 |
CYP11A1 |
cytochrome P450, family 11, subfamily A, polypeptide 1 |
19273 |
0.13 |
| chr17_78933014_78933165 | 0.66 |
CTD-2561B21.3 |
|
2424 |
0.18 |
| chr10_88409177_88409328 | 0.65 |
OPN4 |
opsin 4 |
5062 |
0.22 |
| chr6_37666285_37666474 | 0.63 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
613 |
0.81 |
| chr9_36859555_36859706 | 0.63 |
RP11-344B23.2 |
|
3078 |
0.23 |
| chr14_105445324_105445475 | 0.63 |
AHNAK2 |
AHNAK nucleoprotein 2 |
705 |
0.63 |
| chr4_6272197_6272532 | 0.62 |
WFS1 |
Wolfram syndrome 1 (wolframin) |
787 |
0.74 |
| chr20_31384666_31384857 | 0.62 |
DNMT3B |
DNA (cytosine-5-)-methyltransferase 3 beta |
17103 |
0.16 |
| chr12_124907481_124907632 | 0.61 |
NCOR2 |
nuclear receptor corepressor 2 |
34186 |
0.22 |
| chr6_106590141_106590398 | 0.61 |
RP1-134E15.3 |
|
42254 |
0.16 |
| chr14_99714170_99714435 | 0.61 |
AL109767.1 |
|
14983 |
0.21 |
| chr22_20226986_20227137 | 0.59 |
RTN4R |
reticulon 4 receptor |
4146 |
0.16 |
| chr11_64609797_64609948 | 0.58 |
CDC42BPG |
CDC42 binding protein kinase gamma (DMPK-like) |
2169 |
0.18 |
| chr22_29705214_29705587 | 0.58 |
GAS2L1 |
growth arrest-specific 2 like 1 |
2352 |
0.17 |
| chr4_3439247_3439398 | 0.57 |
HGFAC |
HGF activator |
4292 |
0.21 |
| chr16_70774609_70775132 | 0.56 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
3613 |
0.18 |
| chr1_19338582_19338733 | 0.56 |
IFFO2 |
intermediate filament family orphan 2 |
55477 |
0.11 |
| chr11_3109293_3109444 | 0.55 |
OSBPL5 |
oxysterol binding protein-like 5 |
6741 |
0.16 |
| chr9_101573597_101573748 | 0.55 |
GALNT12 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) |
3691 |
0.24 |
| chr10_95229262_95229590 | 0.55 |
MYOF |
myoferlin |
12525 |
0.19 |
| chr1_193155079_193155230 | 0.54 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
630 |
0.75 |
| chr20_60945069_60945220 | 0.53 |
LAMA5 |
laminin, alpha 5 |
2776 |
0.17 |
| chr9_131675686_131675837 | 0.53 |
PHYHD1 |
phytanoyl-CoA dioxygenase domain containing 1 |
7413 |
0.12 |
| chr8_124553954_124554105 | 0.53 |
FBXO32 |
F-box protein 32 |
583 |
0.77 |
| chr5_175257162_175257313 | 0.52 |
CPLX2 |
complexin 2 |
31394 |
0.18 |
| chr3_55525759_55525910 | 0.52 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1861 |
0.41 |
| chr18_77937535_77937686 | 0.52 |
AC139100.3 |
|
3953 |
0.23 |
| chr10_5567726_5567877 | 0.52 |
RP11-116G8.5 |
|
96 |
0.8 |
| chr3_128016629_128016780 | 0.51 |
ENSG00000221067 |
. |
64304 |
0.11 |
| chr9_140507478_140507731 | 0.51 |
C9orf37 |
chromosome 9 open reading frame 37 |
5661 |
0.12 |
| chr10_5605742_5605953 | 0.51 |
CALML3-AS1 |
CALML3 antisense RNA 1 |
37638 |
0.11 |
| chr12_123555934_123556085 | 0.50 |
PITPNM2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
4633 |
0.2 |
| chr21_45715243_45715525 | 0.50 |
PFKL |
phosphofructokinase, liver |
4550 |
0.11 |
| chr1_6419961_6420398 | 0.50 |
ACOT7 |
acyl-CoA thioesterase 7 |
587 |
0.69 |
| chr15_45455194_45455384 | 0.50 |
RP11-519G16.2 |
|
7253 |
0.1 |
| chr16_85388240_85388391 | 0.49 |
RP11-680G10.1 |
Uncharacterized protein |
2754 |
0.33 |
| chr12_94441922_94442073 | 0.48 |
ENSG00000223126 |
. |
39174 |
0.18 |
| chr18_47205073_47205224 | 0.48 |
LIPG |
lipase, endothelial |
116433 |
0.04 |
| chr14_68935288_68935439 | 0.48 |
RAD51B |
RAD51 paralog B |
57136 |
0.15 |
| chr3_159505193_159505344 | 0.48 |
IQCJ-SCHIP1-AS1 |
IQCJ-SCHIP1 readthrough antisense RNA 1 |
18867 |
0.15 |
| chr12_52546716_52546867 | 0.47 |
KRT80 |
keratin 80 |
38993 |
0.08 |
| chr15_74472280_74472489 | 0.47 |
RP11-665J16.1 |
|
817 |
0.48 |
| chr4_6274602_6274753 | 0.47 |
WFS1 |
Wolfram syndrome 1 (wolframin) |
3100 |
0.35 |
| chr7_130003657_130003904 | 0.46 |
CPA5 |
carboxypeptidase A5 |
498 |
0.72 |
| chr14_102172572_102172843 | 0.46 |
RP11-796G6.2 |
Uncharacterized protein |
24067 |
0.17 |
| chr14_32414488_32414678 | 0.46 |
RP11-187E13.1 |
Uncharacterized protein |
524 |
0.84 |
| chr6_116772654_116772805 | 0.46 |
ENSG00000265516 |
. |
6256 |
0.14 |
| chr4_2801177_2801328 | 0.46 |
SH3BP2 |
SH3-domain binding protein 2 |
527 |
0.8 |
| chr7_1562610_1562761 | 0.45 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
7665 |
0.14 |
| chr7_73120336_73120487 | 0.45 |
ENSG00000265724 |
. |
5236 |
0.11 |
| chr12_123429411_123429620 | 0.45 |
ABCB9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
293 |
0.88 |
| chr1_26217938_26218089 | 0.45 |
STMN1 |
stathmin 1 |
13606 |
0.08 |
| chr3_42556058_42556209 | 0.45 |
VIPR1 |
vasoactive intestinal peptide receptor 1 |
12018 |
0.15 |
| chr11_118492770_118493048 | 0.45 |
AP002954.3 |
|
14429 |
0.11 |
| chr16_57831186_57831470 | 0.45 |
KIFC3 |
kinesin family member C3 |
348 |
0.84 |
| chr2_87842171_87842497 | 0.44 |
RP11-1399P15.1 |
|
64781 |
0.13 |
| chr11_94480509_94480660 | 0.44 |
RP11-867G2.8 |
|
7063 |
0.23 |
| chr14_23011383_23011534 | 0.44 |
TRAJ15 |
T cell receptor alpha joining 15 |
12878 |
0.1 |
| chr2_73297957_73298126 | 0.44 |
SFXN5 |
sideroflexin 5 |
761 |
0.67 |
| chr17_38600137_38600500 | 0.44 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
605 |
0.66 |
| chr7_966993_967144 | 0.43 |
ADAP1 |
ArfGAP with dual PH domains 1 |
82 |
0.96 |
| chr19_4584744_4584943 | 0.43 |
SEMA6B |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
25023 |
0.09 |
| chr14_104321166_104321317 | 0.43 |
PPP1R13B |
protein phosphatase 1, regulatory subunit 13B |
7314 |
0.17 |
| chr14_100040999_100041176 | 0.43 |
CCDC85C |
coiled-coil domain containing 85C |
1775 |
0.33 |
| chr19_49119290_49119441 | 0.43 |
FAM83E |
family with sequence similarity 83, member E |
1293 |
0.21 |
| chr1_43415164_43415399 | 0.42 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
9219 |
0.19 |
| chr12_3228682_3229088 | 0.42 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
30078 |
0.18 |
| chr7_150635354_150635936 | 0.42 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
17349 |
0.15 |
| chr2_113392435_113392586 | 0.42 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
10924 |
0.19 |
| chr4_6274786_6274937 | 0.42 |
WFS1 |
Wolfram syndrome 1 (wolframin) |
3284 |
0.34 |
| chr2_97561253_97561466 | 0.42 |
FAM178B |
family with sequence similarity 178, member B |
2511 |
0.22 |
| chr16_68278720_68278897 | 0.42 |
PLA2G15 |
phospholipase A2, group XV |
399 |
0.66 |
| chr17_41938860_41939011 | 0.42 |
CD300LG |
CD300 molecule-like family member g |
14391 |
0.11 |
| chr1_8255073_8255224 | 0.42 |
ENSG00000200975 |
. |
11509 |
0.23 |
| chr2_111367449_111367600 | 0.42 |
AC123886.2 |
|
969 |
0.61 |
| chr2_219472098_219472249 | 0.42 |
PLCD4 |
phospholipase C, delta 4 |
315 |
0.8 |
| chr7_99760518_99760936 | 0.42 |
GAL3ST4 |
galactose-3-O-sulfotransferase 4 |
4126 |
0.07 |
| chr2_98964621_98964860 | 0.42 |
AC092675.4 |
|
1282 |
0.37 |
| chr17_79856034_79856185 | 0.41 |
NPB |
neuropeptide B |
2512 |
0.09 |
| chr22_20229537_20229756 | 0.41 |
RTN4R |
reticulon 4 receptor |
1561 |
0.29 |
| chr16_51189445_51189596 | 0.41 |
SALL1 |
spalt-like transcription factor 1 |
4334 |
0.27 |
| chr15_84048823_84048974 | 0.41 |
SH3GL3 |
SH3-domain GRB2-like 3 |
67082 |
0.11 |
| chr11_133818315_133818530 | 0.41 |
IGSF9B |
immunoglobulin superfamily, member 9B |
2235 |
0.31 |
| chr21_45502719_45503242 | 0.41 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
24191 |
0.15 |
| chr2_133173738_133173889 | 0.41 |
GPR39 |
G protein-coupled receptor 39 |
334 |
0.9 |
| chr15_76488697_76488848 | 0.41 |
RP11-593F23.1 |
|
14945 |
0.24 |
| chr19_38879548_38879838 | 0.40 |
SPRED3 |
sprouty-related, EVH1 domain containing 3 |
559 |
0.46 |
| chr10_116267845_116267996 | 0.40 |
ABLIM1 |
actin binding LIM protein 1 |
18674 |
0.24 |
| chr1_210405648_210405799 | 0.40 |
SERTAD4 |
SERTA domain containing 4 |
421 |
0.6 |
| chr16_811264_811437 | 0.40 |
MSLN |
mesothelin |
252 |
0.78 |
| chr6_37592095_37592288 | 0.39 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
21959 |
0.22 |
| chr10_80918331_80918482 | 0.39 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
89614 |
0.09 |
| chr5_31988023_31988174 | 0.39 |
ENSG00000266243 |
. |
51833 |
0.13 |
| chr2_235932870_235933072 | 0.39 |
SH3BP4 |
SH3-domain binding protein 4 |
29487 |
0.26 |
| chr2_69534306_69534473 | 0.39 |
ENSG00000252250 |
. |
27343 |
0.2 |
| chr2_28977323_28977503 | 0.39 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
2694 |
0.24 |
| chr12_112195610_112195963 | 0.39 |
RP11-162P23.2 |
|
4092 |
0.21 |
| chr1_47077504_47077655 | 0.38 |
MOB3C |
MOB kinase activator 3C |
3226 |
0.18 |
| chr20_61468104_61468255 | 0.38 |
COL9A3 |
collagen, type IX, alpha 3 |
19016 |
0.11 |
| chr15_80472063_80472219 | 0.38 |
FAH |
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
23848 |
0.2 |
| chr20_60892338_60892489 | 0.38 |
RP11-157P1.4 |
|
11032 |
0.12 |
| chr1_156631459_156631667 | 0.38 |
RP11-284F21.7 |
|
347 |
0.75 |
| chr11_112411093_112411244 | 0.38 |
ENSG00000212397 |
. |
62007 |
0.14 |
| chr6_160399730_160400221 | 0.38 |
IGF2R |
insulin-like growth factor 2 receptor |
9844 |
0.21 |
| chr3_33033448_33033599 | 0.38 |
CCR4 |
chemokine (C-C motif) receptor 4 |
40457 |
0.16 |
| chr13_95898574_95898725 | 0.37 |
ENSG00000238463 |
. |
36051 |
0.17 |
| chr2_110519060_110519211 | 0.37 |
RGPD5 |
RANBP2-like and GRIP domain containing 5 |
31200 |
0.22 |
| chr4_147443936_147444348 | 0.37 |
SLC10A7 |
solute carrier family 10, member 7 |
1019 |
0.64 |
| chr1_19255265_19255716 | 0.37 |
IFFO2 |
intermediate filament family orphan 2 |
17004 |
0.16 |
| chr7_578321_578511 | 0.37 |
ENSG00000252986 |
. |
11954 |
0.17 |
| chr11_66137353_66137504 | 0.37 |
SLC29A2 |
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
1863 |
0.14 |
| chr17_79386944_79387260 | 0.37 |
ENSG00000266392 |
. |
12524 |
0.11 |
| chr2_238887461_238887612 | 0.37 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
5799 |
0.22 |
| chr21_33894297_33894448 | 0.37 |
ENSG00000252045 |
. |
16237 |
0.16 |
| chr5_176869820_176870094 | 0.36 |
PRR7-AS1 |
PRR7 antisense RNA 1 |
1521 |
0.22 |
| chr16_57849254_57849515 | 0.36 |
CTD-2600O9.1 |
uncharacterized protein LOC388282 |
1700 |
0.28 |
| chr16_30081154_30081305 | 0.36 |
ALDOA |
aldolase A, fructose-bisphosphate |
330 |
0.73 |
| chr19_50289714_50289865 | 0.36 |
AP2A1 |
adaptor-related protein complex 2, alpha 1 subunit |
19398 |
0.06 |
| chr15_99983240_99983408 | 0.36 |
MEF2A |
myocyte enhancer factor 2A |
34046 |
0.15 |
| chr2_120156492_120156643 | 0.36 |
TMEM37 |
transmembrane protein 37 |
30910 |
0.14 |
| chr8_141599068_141599368 | 0.36 |
AGO2 |
argonaute RISC catalytic component 2 |
16769 |
0.22 |
| chr1_27668239_27668429 | 0.36 |
SYTL1 |
synaptotagmin-like 1 |
179 |
0.91 |
| chr17_26832603_26832754 | 0.36 |
FOXN1 |
forkhead box N1 |
583 |
0.59 |
| chr22_46502171_46502327 | 0.35 |
ENSG00000198986 |
. |
6380 |
0.1 |
| chr19_18596855_18597476 | 0.35 |
ELL |
elongation factor RNA polymerase II |
10422 |
0.1 |
| chr17_4641577_4641728 | 0.35 |
CXCL16 |
chemokine (C-X-C motif) ligand 16 |
788 |
0.36 |
| chr4_182887638_182887789 | 0.35 |
AC108142.1 |
|
118524 |
0.06 |
| chr8_11414885_11415154 | 0.35 |
RP11-148O21.4 |
|
512 |
0.71 |
| chr15_31657045_31657355 | 0.35 |
KLF13 |
Kruppel-like factor 13 |
1157 |
0.65 |
| chr6_54711184_54711335 | 0.35 |
FAM83B |
family with sequence similarity 83, member B |
310 |
0.91 |
| chr15_67400398_67400572 | 0.35 |
SMAD3 |
SMAD family member 3 |
9468 |
0.25 |
| chr12_109006057_109006234 | 0.35 |
TMEM119 |
transmembrane protein 119 |
14049 |
0.12 |
| chr11_3035348_3035499 | 0.35 |
CARS-AS1 |
CARS antisense RNA 1 |
15201 |
0.11 |
| chr5_172754453_172754739 | 0.35 |
STC2 |
stanniocalcin 2 |
460 |
0.82 |
| chr4_55099953_55100185 | 0.34 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
3580 |
0.33 |
| chr15_75250496_75250705 | 0.34 |
RPP25 |
ribonuclease P/MRP 25kDa subunit |
795 |
0.51 |
| chr21_43422027_43422309 | 0.34 |
ZBTB21 |
zinc finger and BTB domain containing 21 |
5998 |
0.21 |
| chr1_2345042_2345250 | 0.34 |
PEX10 |
peroxisomal biogenesis factor 10 |
90 |
0.94 |
| chr22_18916870_18917021 | 0.34 |
PRODH |
proline dehydrogenase (oxidase) 1 |
5396 |
0.18 |
| chr3_182792190_182792488 | 0.34 |
MCCC1 |
methylcrotonoyl-CoA carboxylase 1 (alpha) |
24967 |
0.19 |
| chr17_39496038_39496239 | 0.34 |
KRT33A |
keratin 33A |
10926 |
0.08 |
| chr1_44030870_44031021 | 0.34 |
PTPRF |
protein tyrosine phosphatase, receptor type, F |
20198 |
0.18 |
| chr1_25307860_25308011 | 0.34 |
RUNX3 |
runt-related transcription factor 3 |
16434 |
0.19 |
| chr11_67183978_67184499 | 0.34 |
CARNS1 |
carnosine synthase 1 |
681 |
0.4 |
| chr1_6647040_6647244 | 0.34 |
ZBTB48 |
zinc finger and BTB domain containing 48 |
6994 |
0.12 |
| chr19_30115060_30115211 | 0.34 |
POP4 |
processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) |
17907 |
0.21 |
| chr19_11249778_11250050 | 0.34 |
SPC24 |
SPC24, NDC80 kinetochore complex component |
16523 |
0.12 |
| chr11_2406504_2406655 | 0.34 |
CD81 |
CD81 molecule |
317 |
0.81 |
| chr15_41244464_41244615 | 0.34 |
CHAC1 |
ChaC, cation transport regulator homolog 1 (E. coli) |
621 |
0.64 |
| chr17_80196204_80196816 | 0.34 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
1464 |
0.26 |
| chr14_75039833_75039984 | 0.34 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
38867 |
0.13 |
| chr3_10975044_10975195 | 0.34 |
SLC6A1 |
solute carrier family 6 (neurotransmitter transporter), member 1 |
59291 |
0.13 |
| chr22_46447401_46447609 | 0.34 |
RP6-109B7.5 |
|
1468 |
0.21 |
| chr7_76236455_76236606 | 0.33 |
POMZP3 |
POM121 and ZP3 fusion |
11087 |
0.2 |
| chr14_105437323_105437474 | 0.33 |
AHNAK2 |
AHNAK nucleoprotein 2 |
7296 |
0.16 |
| chr16_69515111_69515262 | 0.33 |
CYB5B |
cytochrome b5 type B (outer mitochondrial membrane) |
56500 |
0.09 |
| chr22_21335373_21335584 | 0.33 |
LZTR1 |
leucine-zipper-like transcription regulator 1 |
824 |
0.39 |
| chr1_2140834_2140985 | 0.33 |
AL590822.1 |
Uncharacterized protein |
4711 |
0.13 |
| chr22_39633799_39634052 | 0.33 |
PDGFB |
platelet-derived growth factor beta polypeptide |
2989 |
0.2 |
| chr10_114394071_114394222 | 0.33 |
ENSG00000264763 |
. |
217 |
0.96 |
| chr2_235406648_235406799 | 0.33 |
ARL4C |
ADP-ribosylation factor-like 4C |
1026 |
0.71 |
| chr16_31141784_31142119 | 0.33 |
RP11-388M20.2 |
|
803 |
0.33 |
| chr12_81471874_81472025 | 0.33 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
113 |
0.98 |
| chr1_20813373_20813524 | 0.33 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
735 |
0.68 |
| chr8_139805893_139806044 | 0.33 |
COL22A1 |
collagen, type XXII, alpha 1 |
14163 |
0.32 |
| chr16_87270556_87270707 | 0.33 |
RP11-899L11.3 |
|
21110 |
0.16 |
| chr17_71820638_71820789 | 0.33 |
SDK2 |
sidekick cell adhesion molecule 2 |
180485 |
0.03 |
| chr6_12325038_12325189 | 0.33 |
EDN1 |
endothelin 1 |
34517 |
0.22 |
| chr17_74526070_74526300 | 0.33 |
CYGB |
cytoglobin |
1962 |
0.16 |
| chr17_39067227_39067378 | 0.32 |
AC004231.2 |
|
10380 |
0.1 |
| chrX_23919381_23919603 | 0.32 |
CXorf58 |
chromosome X open reading frame 58 |
6426 |
0.2 |
| chr7_28398441_28398592 | 0.32 |
CREB5 |
cAMP responsive element binding protein 5 |
50490 |
0.18 |
| chr21_42441104_42441255 | 0.32 |
LINC00323 |
long intergenic non-protein coding RNA 323 |
78171 |
0.09 |
| chr9_114736508_114736659 | 0.32 |
ENSG00000222356 |
. |
22436 |
0.17 |
| chr3_45225003_45225154 | 0.32 |
CDCP1 |
CUB domain containing protein 1 |
37164 |
0.15 |
| chr14_102197077_102197344 | 0.32 |
RP11-796G6.2 |
Uncharacterized protein |
436 |
0.83 |
| chr1_3390429_3390580 | 0.32 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
2316 |
0.31 |
| chr1_185703531_185703682 | 0.32 |
HMCN1 |
hemicentin 1 |
77 |
0.98 |
| chr7_55322085_55322386 | 0.32 |
EGFR-AS1 |
EGFR antisense RNA 1 |
65608 |
0.13 |
| chr8_101428189_101428340 | 0.32 |
ENSG00000265599 |
. |
33273 |
0.16 |
| chr7_29663646_29663797 | 0.32 |
DPY19L2P3 |
DPY19L2 pseudogene 3 |
26122 |
0.18 |
| chr6_147828281_147828543 | 0.32 |
SAMD5 |
sterile alpha motif domain containing 5 |
1651 |
0.55 |
| chr9_137221477_137221979 | 0.32 |
RXRA |
retinoid X receptor, alpha |
3302 |
0.32 |
| chr1_19395472_19395623 | 0.32 |
UBR4 |
ubiquitin protein ligase E3 component n-recognin 4 |
13187 |
0.19 |
| chr17_41439641_41439819 | 0.31 |
ENSG00000188825 |
. |
25055 |
0.11 |
| chr6_35048787_35048938 | 0.31 |
TCP11 |
t-complex 11, testis-specific |
39960 |
0.16 |
| chr10_73869748_73870117 | 0.31 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
21142 |
0.19 |
| chr10_29768661_29768812 | 0.31 |
SVIL |
supervillin |
16711 |
0.22 |
| chr10_73769753_73769963 | 0.31 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
45735 |
0.15 |
| chr10_75494456_75494607 | 0.31 |
GLUD1P3 |
glutamate dehydrogenase 1 pseudogene 3 |
3597 |
0.1 |
| chr22_37809155_37809306 | 0.31 |
RP1-63G5.5 |
|
14250 |
0.14 |
| chr10_33624973_33625145 | 0.31 |
NRP1 |
neuropilin 1 |
131 |
0.98 |
| chr16_15235562_15235795 | 0.31 |
ENSG00000207294 |
. |
8876 |
0.11 |
| chr22_37427155_37427306 | 0.31 |
MPST |
mercaptopyruvate sulfurtransferase |
8152 |
0.1 |
| chr4_1757359_1757510 | 0.31 |
TACC3 |
transforming, acidic coiled-coil containing protein 3 |
27303 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.4 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.6 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.1 | 0.3 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.9 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.2 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.1 | 0.3 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.4 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.1 | 0.2 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.2 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.1 | 0.2 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.4 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.2 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.1 | 0.4 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.1 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.1 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 0.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.2 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.3 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.2 | GO:0060534 | trachea cartilage development(GO:0060534) |
| 0.0 | 0.1 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.2 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.2 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.2 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.3 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:0072162 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.3 | GO:0055093 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.2 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.2 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.1 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.2 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:0046475 | phosphatidylcholine catabolic process(GO:0034638) glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0051583 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.3 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) |
| 0.0 | 0.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.3 | GO:0001738 | morphogenesis of a polarized epithelium(GO:0001738) |
| 0.0 | 0.5 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.0 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0060581 | ventral spinal cord interneuron differentiation(GO:0021514) ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:0071453 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.0 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0031958 | corticosteroid receptor signaling pathway(GO:0031958) glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:0043525 | positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.0 | GO:0010985 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.0 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) lymphocyte migration(GO:0072676) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.2 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.3 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0090003 | regulation of establishment of protein localization to plasma membrane(GO:0090003) regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 | 0.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.0 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.1 | GO:0032096 | negative regulation of response to food(GO:0032096) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.0 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.6 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.0 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0045123 | cellular extravasation(GO:0045123) |
| 0.0 | 0.1 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.1 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.0 | GO:0098810 | neurotransmitter reuptake(GO:0098810) |
| 0.0 | 0.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.0 | GO:0021548 | pons development(GO:0021548) |
| 0.0 | 0.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.2 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0051828 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.0 | GO:0060192 | negative regulation of lipase activity(GO:0060192) |
| 0.0 | 0.0 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.0 | GO:1903019 | negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.0 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 | 0.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.0 | GO:0048302 | regulation of isotype switching to IgG isotypes(GO:0048302) |
| 0.0 | 0.2 | GO:0000080 | mitotic G1 phase(GO:0000080) |
| 0.0 | 0.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.0 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.0 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.1 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.2 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.0 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.4 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.1 | 0.3 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.3 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.3 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.1 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.4 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 1.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0051192 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.2 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.4 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.0 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.0 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |