Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
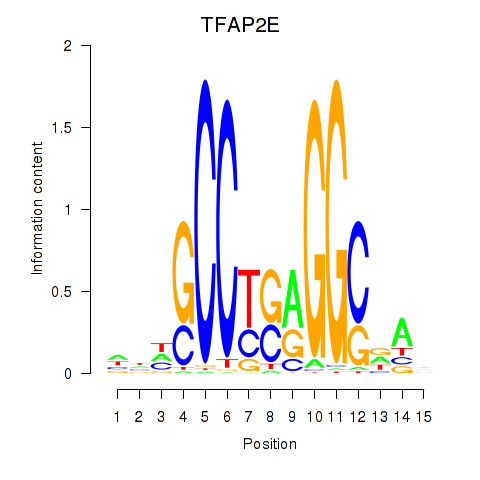
Results for TFAP2E
Z-value: 1.13
Transcription factors associated with TFAP2E
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2E
|
ENSG00000116819.6 | transcription factor AP-2 epsilon |
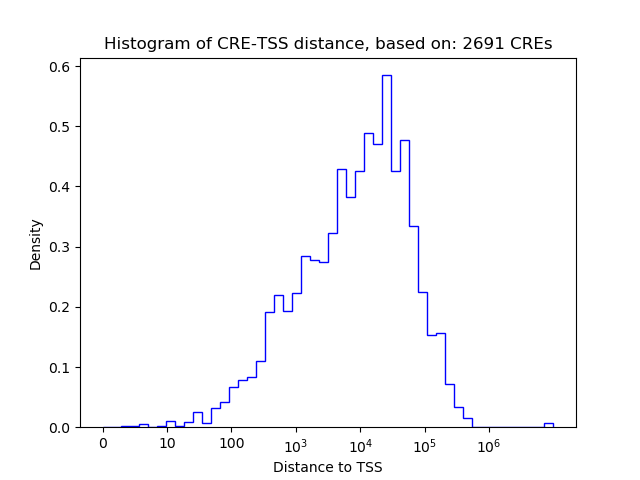
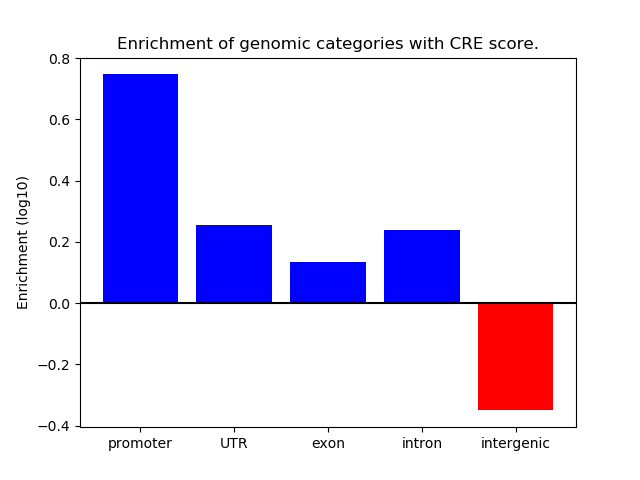
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_36034994_36035145 | TFAP2E | 3902 | 0.163329 | 0.87 | 2.2e-03 | Click! |
| chr1_36035927_36036078 | TFAP2E | 2969 | 0.182736 | 0.70 | 3.5e-02 | Click! |
| chr1_36038348_36038499 | TFAP2E | 548 | 0.566719 | 0.37 | 3.2e-01 | Click! |
| chr1_36035265_36035416 | TFAP2E | 3631 | 0.167671 | 0.37 | 3.3e-01 | Click! |
| chr1_36038695_36039018 | TFAP2E | 115 | 0.844928 | 0.03 | 9.3e-01 | Click! |
Activity of the TFAP2E motif across conditions
Conditions sorted by the z-value of the TFAP2E motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
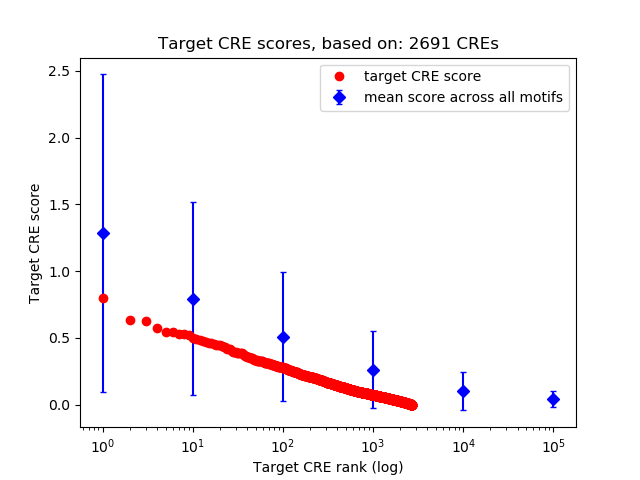
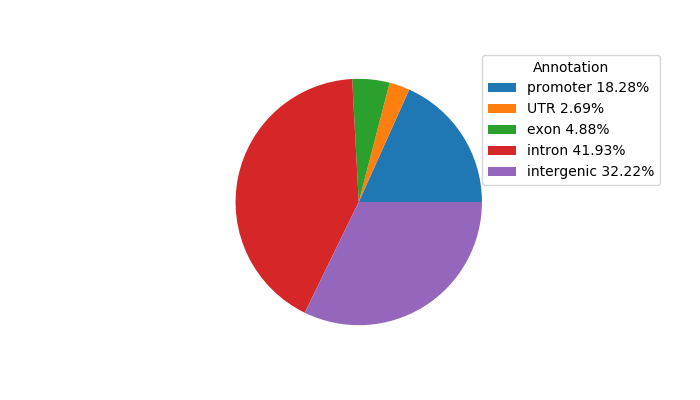
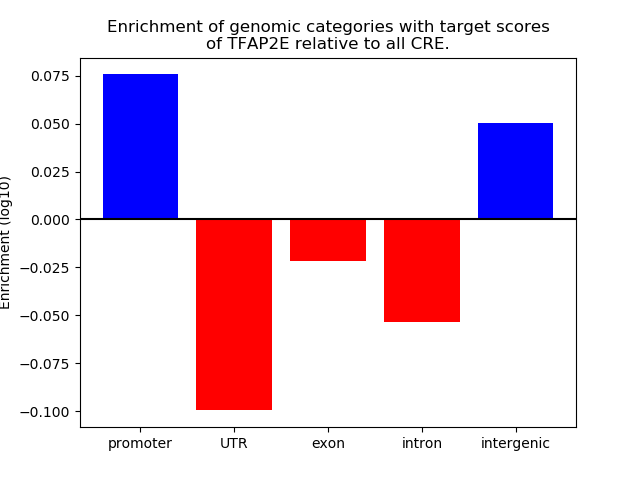
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_127489904_127490214 | 0.80 |
MGLL |
monoglyceride lipase |
12792 |
0.24 |
| chr7_155377735_155377886 | 0.64 |
CNPY1 |
canopy FGF signaling regulator 1 |
51253 |
0.12 |
| chr10_18071932_18072083 | 0.63 |
MRC1 |
mannose receptor, C type 1 |
26345 |
0.17 |
| chr2_238783166_238783317 | 0.57 |
ENSG00000263723 |
. |
4694 |
0.22 |
| chr20_16507696_16507995 | 0.55 |
KIF16B |
kinesin family member 16B |
46233 |
0.18 |
| chr8_96237601_96237983 | 0.54 |
C8orf37 |
chromosome 8 open reading frame 37 |
43637 |
0.17 |
| chr20_33166706_33166857 | 0.53 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
20259 |
0.16 |
| chr10_96974401_96974606 | 0.53 |
RP11-310E22.4 |
|
16398 |
0.21 |
| chr10_104539691_104539842 | 0.52 |
WBP1L |
WW domain binding protein 1-like |
3760 |
0.18 |
| chr2_69034764_69034964 | 0.50 |
ARHGAP25 |
Rho GTPase activating protein 25 |
501 |
0.82 |
| chr8_8269357_8269508 | 0.49 |
SGK223 |
Tyrosine-protein kinase SgK223 |
25424 |
0.23 |
| chr15_85215324_85215475 | 0.49 |
NMB |
neuromedin B |
13605 |
0.1 |
| chr2_9815348_9815499 | 0.48 |
YWHAQ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
44280 |
0.14 |
| chr9_136741627_136741778 | 0.47 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
115701 |
0.05 |
| chr12_122519590_122520270 | 0.47 |
MLXIP |
MLX interacting protein |
3302 |
0.29 |
| chr17_27910417_27910568 | 0.46 |
GIT1 |
G protein-coupled receptor kinase interacting ArfGAP 1 |
398 |
0.68 |
| chr15_44205800_44205951 | 0.46 |
FRMD5 |
FERM domain containing 5 |
5825 |
0.19 |
| chr4_5638816_5638967 | 0.45 |
EVC2 |
Ellis van Creveld syndrome 2 |
71403 |
0.11 |
| chr5_92629860_92630011 | 0.45 |
ENSG00000237187 |
. |
128184 |
0.06 |
| chr2_40662476_40662683 | 0.45 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
5135 |
0.35 |
| chr2_153406171_153406322 | 0.44 |
FMNL2 |
formin-like 2 |
69846 |
0.13 |
| chr1_118644337_118644488 | 0.43 |
SPAG17 |
sperm associated antigen 17 |
83434 |
0.1 |
| chr2_102680238_102680787 | 0.43 |
IL1R1 |
interleukin 1 receptor, type I |
492 |
0.85 |
| chr8_143762185_143762336 | 0.42 |
PSCA |
prostate stem cell antigen |
385 |
0.77 |
| chr15_72487652_72487940 | 0.42 |
GRAMD2 |
GRAM domain containing 2 |
2330 |
0.25 |
| chr14_65703391_65703542 | 0.41 |
ENSG00000266740 |
. |
98435 |
0.07 |
| chr5_81661345_81661496 | 0.40 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
60254 |
0.15 |
| chr19_8053652_8053803 | 0.39 |
ELAVL1 |
ELAV like RNA binding protein 1 |
7731 |
0.11 |
| chr12_32242647_32242798 | 0.39 |
BICD1 |
bicaudal D homolog 1 (Drosophila) |
17047 |
0.21 |
| chr8_124468816_124468967 | 0.39 |
WDYHV1 |
WDYHV motif containing 1 |
26656 |
0.16 |
| chr7_155049692_155049843 | 0.39 |
INSIG1 |
insulin induced gene 1 |
39719 |
0.15 |
| chr2_112255788_112255939 | 0.39 |
ENSG00000266139 |
. |
177195 |
0.03 |
| chr5_110789980_110790131 | 0.39 |
STARD4-AS1 |
STARD4 antisense RNA 1 |
57869 |
0.12 |
| chr1_110598662_110598813 | 0.39 |
RP4-773N10.4 |
|
2225 |
0.21 |
| chr17_47786040_47786409 | 0.38 |
SLC35B1 |
solute carrier family 35, member B1 |
152 |
0.82 |
| chr16_70414605_70414756 | 0.38 |
RP11-529K1.4 |
|
1320 |
0.31 |
| chr3_54925868_54926132 | 0.37 |
CACNA2D3-AS1 |
CACNA2D3 antisense RNA 1 |
9282 |
0.27 |
| chr7_137564480_137564636 | 0.37 |
DGKI |
diacylglycerol kinase, iota |
32720 |
0.18 |
| chr11_27444813_27444964 | 0.37 |
RP11-159H22.2 |
|
48388 |
0.12 |
| chr6_11212735_11212977 | 0.36 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
20035 |
0.2 |
| chr15_67316146_67316685 | 0.36 |
SMAD3 |
SMAD family member 3 |
39686 |
0.2 |
| chr1_211665583_211665734 | 0.35 |
RD3 |
retinal degeneration 3 |
601 |
0.8 |
| chr12_112550183_112550334 | 0.35 |
NAA25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
3432 |
0.18 |
| chr19_14669270_14669558 | 0.35 |
TECR |
trans-2,3-enoyl-CoA reductase |
3365 |
0.13 |
| chr22_38531557_38531708 | 0.35 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
2748 |
0.19 |
| chr19_4712858_4713009 | 0.35 |
DPP9 |
dipeptidyl-peptidase 9 |
1385 |
0.3 |
| chr5_167243493_167243835 | 0.34 |
ENSG00000221741 |
. |
54006 |
0.13 |
| chr2_216783049_216783200 | 0.34 |
ENSG00000212055 |
. |
39482 |
0.19 |
| chr12_3231998_3232272 | 0.34 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
26828 |
0.19 |
| chr4_185739235_185739386 | 0.33 |
RP11-701P16.2 |
Uncharacterized protein |
4537 |
0.2 |
| chr1_179096650_179096926 | 0.33 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
15391 |
0.19 |
| chr11_111794694_111794845 | 0.33 |
CRYAB |
crystallin, alpha B |
323 |
0.78 |
| chr15_31898343_31898494 | 0.33 |
OTUD7A |
OTU domain containing 7A |
49124 |
0.19 |
| chr10_79152930_79153197 | 0.33 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
10679 |
0.24 |
| chr1_17519325_17519476 | 0.33 |
PADI1 |
peptidyl arginine deiminase, type I |
12221 |
0.17 |
| chr4_55529764_55530043 | 0.33 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
5818 |
0.34 |
| chr2_20777848_20777999 | 0.33 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
14385 |
0.21 |
| chr4_142273560_142273938 | 0.33 |
ENSG00000238695 |
. |
54717 |
0.16 |
| chr4_78783004_78783324 | 0.32 |
MRPL1 |
mitochondrial ribosomal protein L1 |
510 |
0.86 |
| chr5_148349011_148349162 | 0.32 |
RP11-44B19.1 |
|
817 |
0.67 |
| chr10_116154509_116154660 | 0.32 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
9655 |
0.23 |
| chr2_164538752_164538903 | 0.32 |
FIGN |
fidgetin |
53690 |
0.19 |
| chr5_9048002_9048358 | 0.32 |
ENSG00000266415 |
. |
5827 |
0.3 |
| chrX_14977641_14977792 | 0.31 |
MOSPD2 |
motile sperm domain containing 2 |
50672 |
0.17 |
| chr17_70461708_70461859 | 0.31 |
ENSG00000200783 |
. |
198508 |
0.03 |
| chr11_124770110_124770261 | 0.31 |
ROBO4 |
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
2008 |
0.19 |
| chr1_161171729_161171880 | 0.31 |
NDUFS2 |
NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) |
133 |
0.88 |
| chr2_159779762_159779913 | 0.31 |
ENSG00000222300 |
. |
12660 |
0.2 |
| chr1_239620290_239620441 | 0.31 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
172008 |
0.04 |
| chr1_115782386_115782537 | 0.31 |
RP4-663N10.1 |
|
43194 |
0.18 |
| chr17_34112154_34112305 | 0.30 |
MMP28 |
matrix metallopeptidase 28 |
5857 |
0.11 |
| chr18_20862079_20862230 | 0.30 |
RP11-17J14.2 |
|
21836 |
0.23 |
| chr1_213293309_213293460 | 0.30 |
RPS6KC1 |
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
68780 |
0.12 |
| chrX_118628293_118628444 | 0.30 |
SLC25A5 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
26005 |
0.12 |
| chr3_149290093_149290244 | 0.30 |
WWTR1 |
WW domain containing transcription regulator 1 |
3871 |
0.25 |
| chr19_13171036_13171187 | 0.30 |
AC007787.2 |
|
11077 |
0.1 |
| chr16_75282266_75282417 | 0.30 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
203 |
0.92 |
| chr20_39122912_39123063 | 0.30 |
ENSG00000252434 |
. |
77472 |
0.12 |
| chr6_14716899_14717050 | 0.30 |
ENSG00000206960 |
. |
70208 |
0.14 |
| chr8_126668851_126669002 | 0.30 |
ENSG00000266452 |
. |
212119 |
0.02 |
| chr1_24691365_24691516 | 0.29 |
STPG1 |
sperm-tail PG-rich repeat containing 1 |
14873 |
0.16 |
| chr11_78494593_78494744 | 0.29 |
TENM4 |
teneurin transmembrane protein 4 |
21788 |
0.22 |
| chr1_161052827_161053090 | 0.29 |
RP11-544M22.8 |
|
1297 |
0.22 |
| chr11_119189693_119189895 | 0.29 |
MCAM |
melanoma cell adhesion molecule |
1968 |
0.16 |
| chr1_45990852_45991003 | 0.29 |
PRDX1 |
peroxiredoxin 1 |
2208 |
0.22 |
| chr6_140783983_140784266 | 0.29 |
ENSG00000221336 |
. |
197726 |
0.03 |
| chr8_41009602_41009753 | 0.29 |
ENSG00000263372 |
. |
118985 |
0.05 |
| chr10_112158236_112158389 | 0.29 |
ENSG00000221359 |
. |
35687 |
0.17 |
| chr12_120667456_120667944 | 0.29 |
PXN |
paxillin |
3053 |
0.16 |
| chr5_77881787_77882057 | 0.29 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
36948 |
0.23 |
| chr17_58218497_58218710 | 0.29 |
CA4 |
carbonic anhydrase IV |
8694 |
0.18 |
| chr1_41356534_41356685 | 0.28 |
RP5-1066H13.4 |
|
27023 |
0.15 |
| chr11_7913917_7914068 | 0.28 |
ENSG00000252769 |
. |
3166 |
0.19 |
| chr3_183740027_183740178 | 0.28 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
4299 |
0.14 |
| chr11_47560816_47560967 | 0.28 |
CELF1 |
CUGBP, Elav-like family member 1 |
13778 |
0.08 |
| chr6_100677123_100677274 | 0.28 |
RP1-121G13.2 |
|
197796 |
0.03 |
| chr2_171829322_171830003 | 0.28 |
GORASP2 |
golgi reassembly stacking protein 2, 55kDa |
43831 |
0.15 |
| chr2_65214115_65214266 | 0.28 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
1434 |
0.38 |
| chr2_110087186_110087362 | 0.28 |
ENSG00000265965 |
. |
157193 |
0.04 |
| chr9_75169117_75169268 | 0.28 |
ENSG00000238402 |
. |
27040 |
0.21 |
| chr2_48646919_48647208 | 0.28 |
PPP1R21 |
protein phosphatase 1, regulatory subunit 21 |
20845 |
0.21 |
| chr10_25348795_25348946 | 0.28 |
ENSG00000266069 |
. |
31613 |
0.16 |
| chr11_44586009_44586190 | 0.28 |
CD82 |
CD82 molecule |
122 |
0.97 |
| chr1_20930163_20930437 | 0.28 |
CDA |
cytidine deaminase |
14859 |
0.16 |
| chr4_38103387_38103538 | 0.27 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
55970 |
0.16 |
| chr1_172446166_172446317 | 0.27 |
C1orf105 |
chromosome 1 open reading frame 105 |
24208 |
0.17 |
| chr10_126029743_126029894 | 0.27 |
OAT |
ornithine aminotransferase |
77687 |
0.1 |
| chr15_62910288_62910439 | 0.27 |
RP11-625H11.1 |
Uncharacterized protein |
27017 |
0.18 |
| chr10_73620641_73620884 | 0.27 |
PSAP |
prosaposin |
9636 |
0.2 |
| chr16_73098361_73099003 | 0.27 |
ZFHX3 |
zinc finger homeobox 3 |
5085 |
0.26 |
| chr17_19215367_19215621 | 0.27 |
EPN2-AS1 |
EPN2 antisense RNA 1 |
5920 |
0.12 |
| chr15_101652404_101652555 | 0.27 |
RP11-505E24.2 |
|
26208 |
0.2 |
| chr8_81665342_81665642 | 0.26 |
ENSG00000212429 |
. |
3564 |
0.28 |
| chr1_62146544_62146695 | 0.26 |
TM2D1 |
TM2 domain containing 1 |
44186 |
0.15 |
| chr20_2795102_2795253 | 0.26 |
TMEM239 |
CDNA FLJ26142 fis, clone TST04526; Transmembrane protein 239; Uncharacterized protein |
437 |
0.45 |
| chr1_53977593_53977891 | 0.26 |
DMRTB1 |
DMRT-like family B with proline-rich C-terminal, 1 |
52670 |
0.14 |
| chr8_96111972_96112794 | 0.26 |
RP11-320N21.2 |
|
12231 |
0.16 |
| chr1_22623912_22624063 | 0.26 |
ENSG00000266564 |
. |
31255 |
0.21 |
| chr12_45829168_45829319 | 0.26 |
ENSG00000239178 |
. |
119199 |
0.06 |
| chr10_126805401_126805552 | 0.26 |
CTBP2 |
C-terminal binding protein 2 |
41809 |
0.18 |
| chr1_167127568_167127862 | 0.26 |
RP11-277B15.2 |
|
37002 |
0.13 |
| chr5_122765001_122765271 | 0.26 |
CEP120 |
centrosomal protein 120kDa |
5850 |
0.29 |
| chr7_150097245_150097396 | 0.25 |
RP4-584D14.6 |
|
16347 |
0.11 |
| chr12_132347924_132348141 | 0.25 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
18192 |
0.15 |
| chr9_129110024_129110380 | 0.25 |
MVB12B |
multivesicular body subunit 12B |
12676 |
0.24 |
| chr7_102515337_102515656 | 0.25 |
ENSG00000238324 |
. |
17599 |
0.16 |
| chr22_36720785_36721049 | 0.25 |
ENSG00000266345 |
. |
8671 |
0.19 |
| chr2_26981199_26981390 | 0.25 |
SLC35F6 |
solute carrier family 35, member F6 |
5862 |
0.19 |
| chr6_125203361_125203883 | 0.25 |
RNF217 |
ring finger protein 217 |
80069 |
0.11 |
| chr16_67742664_67742903 | 0.25 |
GFOD2 |
glucose-fructose oxidoreductase domain containing 2 |
10541 |
0.11 |
| chr16_68986638_68986789 | 0.25 |
RP11-521L9.2 |
|
18990 |
0.18 |
| chr3_128267799_128268165 | 0.25 |
C3orf27 |
chromosome 3 open reading frame 27 |
26947 |
0.16 |
| chrX_45182411_45182562 | 0.25 |
RP11-342D14.1 |
|
2783 |
0.41 |
| chr1_32049799_32049950 | 0.25 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
7735 |
0.14 |
| chr19_42900464_42900801 | 0.25 |
LIPE-AS1 |
LIPE antisense RNA 1 |
648 |
0.56 |
| chr7_76057799_76057950 | 0.25 |
ZP3 |
zona pellucida glycoprotein 3 (sperm receptor) |
81 |
0.96 |
| chr10_116072655_116072806 | 0.25 |
VWA2 |
von Willebrand factor A domain containing 2 |
73641 |
0.09 |
| chr20_48310267_48310418 | 0.25 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
20073 |
0.18 |
| chr2_201622113_201622264 | 0.25 |
ENSG00000201737 |
. |
18119 |
0.14 |
| chr17_32698121_32698371 | 0.24 |
CCL1 |
chemokine (C-C motif) ligand 1 |
7996 |
0.18 |
| chr4_5637335_5637511 | 0.24 |
EVC2 |
Ellis van Creveld syndrome 2 |
72871 |
0.1 |
| chr5_81701965_81702116 | 0.24 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
100874 |
0.08 |
| chr2_145082101_145082252 | 0.24 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
4739 |
0.34 |
| chr7_127661423_127661614 | 0.24 |
LRRC4 |
leucine rich repeat containing 4 |
9540 |
0.25 |
| chr8_37468333_37468484 | 0.24 |
ENSG00000223215 |
. |
58008 |
0.1 |
| chr14_23316976_23317711 | 0.24 |
ENSG00000212335 |
. |
4594 |
0.09 |
| chr1_184591672_184591823 | 0.24 |
ENSG00000252790 |
. |
55321 |
0.15 |
| chr17_66449042_66449331 | 0.24 |
WIPI1 |
WD repeat domain, phosphoinositide interacting 1 |
4376 |
0.23 |
| chr13_33804052_33804203 | 0.23 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
23984 |
0.2 |
| chr20_30199424_30199575 | 0.23 |
ENSG00000264395 |
. |
4510 |
0.13 |
| chrX_22153582_22153733 | 0.23 |
PHEX-AS1 |
PHEX antisense RNA 1 |
37443 |
0.16 |
| chr14_59219843_59220151 | 0.23 |
ENSG00000221427 |
. |
17967 |
0.27 |
| chr15_74274351_74275014 | 0.23 |
STOML1 |
stomatin (EPB72)-like 1 |
9493 |
0.15 |
| chr13_32708450_32708601 | 0.23 |
FRY |
furry homolog (Drosophila) |
73524 |
0.11 |
| chr13_49671478_49671629 | 0.23 |
ENSG00000199788 |
. |
2026 |
0.36 |
| chr2_28660054_28660205 | 0.23 |
PLB1 |
phospholipase B1 |
19883 |
0.17 |
| chr8_26498187_26498401 | 0.23 |
DPYSL2 |
dihydropyrimidinase-like 2 |
62373 |
0.14 |
| chr5_125808253_125808404 | 0.23 |
RP11-517I3.1 |
|
5005 |
0.23 |
| chr2_33352313_33352464 | 0.23 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
7085 |
0.3 |
| chr19_39853657_39853808 | 0.22 |
ENSG00000241983 |
. |
6061 |
0.09 |
| chr22_35907056_35907207 | 0.22 |
RASD2 |
RASD family, member 2 |
29784 |
0.19 |
| chr18_21795564_21795715 | 0.22 |
ENSG00000206766 |
. |
2625 |
0.23 |
| chr6_167142675_167142845 | 0.22 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
28639 |
0.23 |
| chr4_139531486_139531637 | 0.22 |
ENSG00000238971 |
. |
201359 |
0.03 |
| chr5_139721285_139721505 | 0.22 |
HBEGF |
heparin-binding EGF-like growth factor |
4793 |
0.14 |
| chr3_69439182_69439463 | 0.22 |
FRMD4B |
FERM domain containing 4B |
3892 |
0.36 |
| chr1_223308438_223308608 | 0.22 |
TLR5 |
toll-like receptor 5 |
425 |
0.91 |
| chr1_230850007_230850201 | 0.22 |
AGT |
angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
61 |
0.97 |
| chr15_91285398_91285560 | 0.22 |
ENSG00000200677 |
. |
13063 |
0.14 |
| chr5_139621230_139621381 | 0.22 |
CTB-131B5.2 |
|
41557 |
0.1 |
| chr14_72180385_72180536 | 0.22 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
94987 |
0.09 |
| chr8_37376201_37376352 | 0.22 |
RP11-150O12.6 |
|
1737 |
0.48 |
| chr3_151893804_151894059 | 0.22 |
MBNL1 |
muscleblind-like splicing regulator 1 |
91898 |
0.08 |
| chr7_134893432_134893608 | 0.22 |
WDR91 |
WD repeat domain 91 |
2131 |
0.24 |
| chr1_144085227_144085501 | 0.22 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
61444 |
0.14 |
| chr6_140784427_140784578 | 0.22 |
ENSG00000221336 |
. |
197348 |
0.03 |
| chr2_85933045_85933196 | 0.22 |
GNLY |
granulysin |
10629 |
0.13 |
| chr22_46498265_46498416 | 0.22 |
ENSG00000198986 |
. |
10289 |
0.1 |
| chr3_15672619_15672770 | 0.22 |
BTD |
biotinidase |
29218 |
0.14 |
| chr1_1295387_1295783 | 0.22 |
MXRA8 |
matrix-remodelling associated 8 |
1572 |
0.16 |
| chr15_35330956_35331107 | 0.22 |
ZNF770 |
zinc finger protein 770 |
50543 |
0.14 |
| chr22_46485560_46485711 | 0.22 |
ENSG00000266533 |
. |
1289 |
0.27 |
| chr14_63904786_63905352 | 0.22 |
ENSG00000264995 |
. |
24655 |
0.16 |
| chr1_42641010_42641161 | 0.22 |
GUCA2A |
guanylate cyclase activator 2A (guanylin) |
10696 |
0.19 |
| chr13_80657315_80657466 | 0.22 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
256404 |
0.02 |
| chr5_142280838_142280989 | 0.22 |
ARHGAP26 |
Rho GTPase activating protein 26 |
5974 |
0.27 |
| chr2_241860537_241860698 | 0.21 |
AC104809.3 |
Protein LOC728763 |
1369 |
0.36 |
| chr10_95136688_95136839 | 0.21 |
MYOF |
myoferlin |
105188 |
0.06 |
| chr8_85095605_85095756 | 0.21 |
RALYL |
RALY RNA binding protein-like |
89 |
0.99 |
| chr22_46487788_46487939 | 0.21 |
ENSG00000266533 |
. |
939 |
0.38 |
| chr2_19015414_19015565 | 0.21 |
NT5C1B |
5'-nucleotidase, cytosolic IB |
244651 |
0.02 |
| chr10_128777765_128777916 | 0.21 |
RP11-223P11.3 |
|
46846 |
0.16 |
| chr17_36433052_36433525 | 0.21 |
RP11-1407O15.2 |
TBC1 domain family member 3 |
11447 |
0.15 |
| chr3_138187920_138188071 | 0.21 |
CEP70 |
centrosomal protein 70kDa |
31027 |
0.18 |
| chr1_215843470_215843621 | 0.21 |
ENSG00000202498 |
. |
40086 |
0.2 |
| chr11_45067696_45067847 | 0.21 |
PRDM11 |
PR domain containing 11 |
47793 |
0.17 |
| chr9_133323468_133323619 | 0.21 |
ENSG00000238298 |
. |
1698 |
0.3 |
| chr12_57611386_57611537 | 0.21 |
NXPH4 |
neurexophilin 4 |
883 |
0.39 |
| chrX_69394860_69395105 | 0.21 |
DGAT2L6 |
diacylglycerol O-acyltransferase 2-like 6 |
2351 |
0.2 |
| chr9_139492984_139493135 | 0.21 |
ENSG00000252440 |
. |
3908 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.1 | 0.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.2 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.1 | 0.2 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.1 | 0.2 | GO:0052305 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.1 | 0.2 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0002686 | negative regulation of leukocyte migration(GO:0002686) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0046471 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.0 | GO:1903579 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.0 | 0.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.1 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.1 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.0 | 0.1 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.1 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0009188 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway(GO:0033145) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.0 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 0.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.0 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.0 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0033646 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 1.0 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.3 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |