Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
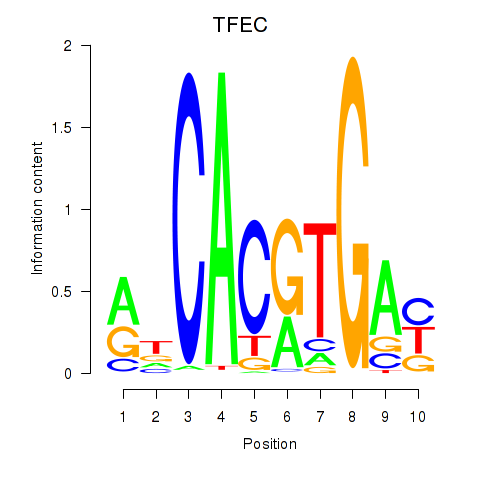
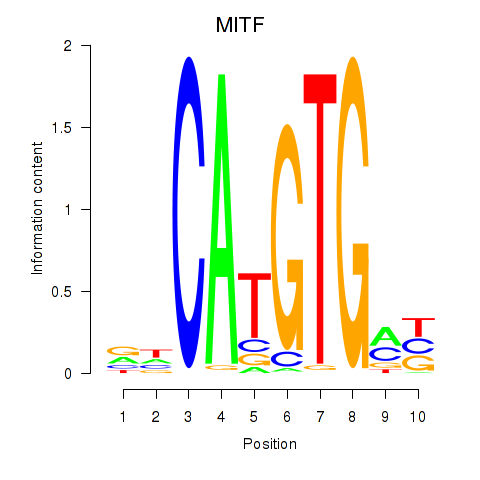
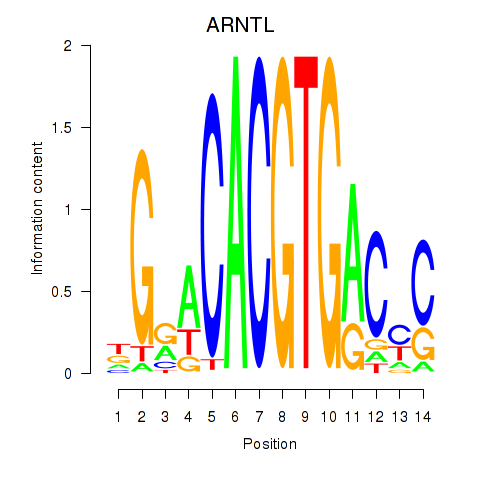
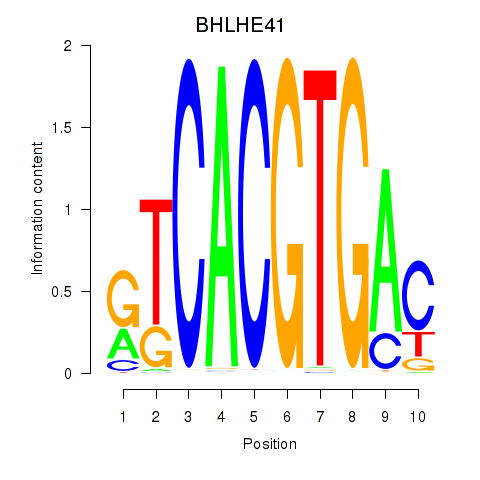
Results for TFEC_MITF_ARNTL_BHLHE41
Z-value: 0.83
Transcription factors associated with TFEC_MITF_ARNTL_BHLHE41
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFEC
|
ENSG00000105967.11 | transcription factor EC |
|
MITF
|
ENSG00000187098.10 | melanocyte inducing transcription factor |
|
ARNTL
|
ENSG00000133794.13 | aryl hydrocarbon receptor nuclear translocator like |
|
BHLHE41
|
ENSG00000123095.5 | basic helix-loop-helix family member e41 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_13230039_13230195 | ARNTL | 68082 | 0.127970 | 0.86 | 3.1e-03 | Click! |
| chr11_13312870_13313021 | ARNTL | 13513 | 0.259982 | -0.83 | 5.8e-03 | Click! |
| chr11_13229796_13230015 | ARNTL | 68294 | 0.127528 | 0.78 | 1.3e-02 | Click! |
| chr11_13282555_13282706 | ARNTL | 15569 | 0.269293 | 0.75 | 2.1e-02 | Click! |
| chr11_13314702_13314853 | ARNTL | 15345 | 0.254012 | -0.73 | 2.5e-02 | Click! |
| chr12_26277295_26278057 | BHLHE41 | 384 | 0.777876 | -0.66 | 5.3e-02 | Click! |
| chr12_26278311_26278658 | BHLHE41 | 424 | 0.539953 | 0.03 | 9.3e-01 | Click! |
| chr3_69789658_69789810 | MITF | 1112 | 0.607008 | 0.80 | 1.0e-02 | Click! |
| chr3_69813958_69814283 | MITF | 1158 | 0.558041 | 0.73 | 2.6e-02 | Click! |
| chr3_69813399_69813761 | MITF | 618 | 0.782625 | 0.70 | 3.4e-02 | Click! |
| chr3_69810828_69810979 | MITF | 972 | 0.633922 | 0.69 | 4.0e-02 | Click! |
| chr3_69788296_69788550 | MITF | 163 | 0.969196 | -0.64 | 6.5e-02 | Click! |
| chr7_115556505_115556656 | TFEC | 51724 | 0.167210 | -0.63 | 7.2e-02 | Click! |
| chr7_115662328_115662479 | TFEC | 8392 | 0.309240 | -0.61 | 8.1e-02 | Click! |
| chr7_115803556_115803707 | TFEC | 3681 | 0.298289 | 0.60 | 8.5e-02 | Click! |
| chr7_115808920_115809071 | TFEC | 9045 | 0.241313 | 0.60 | 8.9e-02 | Click! |
| chr7_115785682_115785833 | TFEC | 14193 | 0.249470 | 0.58 | 1.0e-01 | Click! |
Activity of the TFEC_MITF_ARNTL_BHLHE41 motif across conditions
Conditions sorted by the z-value of the TFEC_MITF_ARNTL_BHLHE41 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
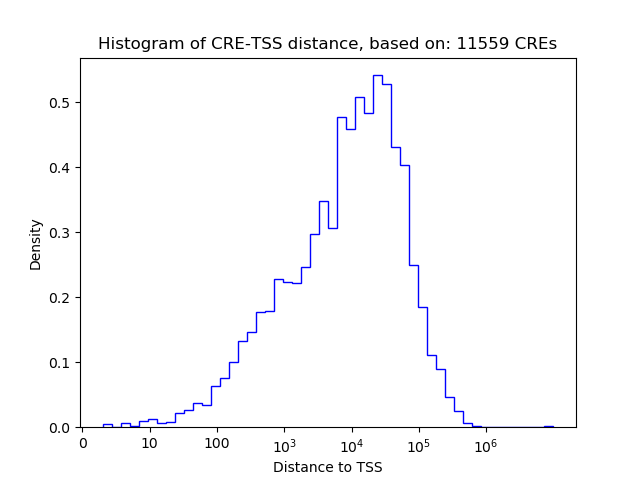
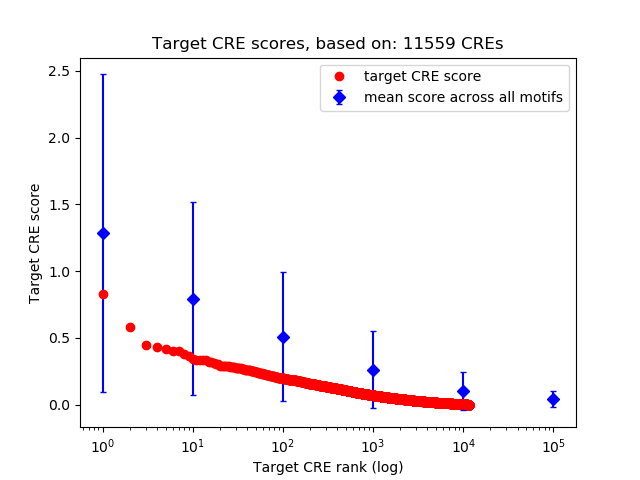
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_114280008_114280159 | 0.83 |
ATP4B |
ATPase, H+/K+ exchanging, beta polypeptide |
32418 |
0.15 |
| chr13_114280229_114280380 | 0.59 |
ATP4B |
ATPase, H+/K+ exchanging, beta polypeptide |
32197 |
0.15 |
| chr16_85549609_85549893 | 0.45 |
ENSG00000264203 |
. |
74653 |
0.09 |
| chr13_44730147_44730391 | 0.43 |
SMIM2-IT1 |
SMIM2 intronic transcript 1 (non-protein coding) |
2089 |
0.32 |
| chr6_16681395_16681795 | 0.41 |
RP1-151F17.1 |
|
79774 |
0.1 |
| chr3_125322024_125322175 | 0.41 |
OSBPL11 |
oxysterol binding protein-like 11 |
8165 |
0.22 |
| chr5_343224_343631 | 0.41 |
AHRR |
aryl-hydrocarbon receptor repressor |
216 |
0.94 |
| chr10_81160624_81160775 | 0.38 |
RP11-342M3.5 |
|
18615 |
0.19 |
| chr2_235866357_235866729 | 0.36 |
SH3BP4 |
SH3-domain binding protein 4 |
5823 |
0.35 |
| chr2_40266109_40266315 | 0.34 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
3278 |
0.4 |
| chr5_14261499_14261650 | 0.34 |
TRIO |
trio Rho guanine nucleotide exchange factor |
29512 |
0.26 |
| chr1_210431394_210431545 | 0.34 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
24077 |
0.19 |
| chr5_14174403_14174686 | 0.33 |
TRIO |
trio Rho guanine nucleotide exchange factor |
9363 |
0.32 |
| chr6_52376693_52376847 | 0.33 |
TRAM2 |
translocation associated membrane protein 2 |
64943 |
0.11 |
| chr18_52454307_52454540 | 0.32 |
RAB27B |
RAB27B, member RAS oncogene family |
41007 |
0.2 |
| chr1_27855366_27855678 | 0.32 |
RP1-159A19.4 |
|
3206 |
0.23 |
| chr14_36538512_36538663 | 0.31 |
ENSG00000212071 |
. |
127187 |
0.05 |
| chr13_40105934_40106139 | 0.31 |
LHFP |
lipoma HMGIC fusion partner |
71272 |
0.11 |
| chr11_10696838_10697077 | 0.30 |
MRVI1 |
murine retrovirus integration site 1 homolog |
18104 |
0.17 |
| chr17_40128851_40129002 | 0.29 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
5413 |
0.13 |
| chr9_82104147_82104538 | 0.29 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
82346 |
0.12 |
| chr21_43953040_43953239 | 0.29 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
18505 |
0.13 |
| chr16_80755255_80755539 | 0.29 |
CTD-2055G21.1 |
|
14860 |
0.21 |
| chr2_9375733_9375964 | 0.29 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
28954 |
0.23 |
| chr17_74526963_74527181 | 0.29 |
CYGB |
cytoglobin |
1075 |
0.29 |
| chr17_3545041_3545192 | 0.29 |
CTNS |
cystinosin, lysosomal cystine transporter |
1675 |
0.21 |
| chr2_47313454_47313781 | 0.28 |
AC073283.7 |
|
18656 |
0.18 |
| chr9_93880537_93881196 | 0.28 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
243311 |
0.02 |
| chr17_9160012_9160446 | 0.28 |
RP11-85B7.4 |
|
26262 |
0.21 |
| chr11_69263417_69264003 | 0.28 |
CCND1 |
cyclin D1 |
192145 |
0.03 |
| chr10_129706485_129706650 | 0.28 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
1151 |
0.53 |
| chr7_104614373_104614618 | 0.28 |
ENSG00000251911 |
. |
2761 |
0.25 |
| chr6_89744917_89745362 | 0.27 |
ENSG00000223001 |
. |
28270 |
0.13 |
| chr15_35598915_35599148 | 0.27 |
ENSG00000265102 |
. |
65534 |
0.14 |
| chr9_137537253_137537404 | 0.27 |
COL5A1 |
collagen, type V, alpha 1 |
3708 |
0.25 |
| chr6_111886228_111886379 | 0.27 |
TRAF3IP2-AS1 |
TRAF3IP2 antisense RNA 1 |
151 |
0.95 |
| chr14_105216209_105216503 | 0.27 |
SIVA1 |
SIVA1, apoptosis-inducing factor |
3081 |
0.16 |
| chr1_9952782_9953028 | 0.26 |
CTNNBIP1 |
catenin, beta interacting protein 1 |
390 |
0.81 |
| chr2_33516894_33517045 | 0.26 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
254 |
0.93 |
| chr1_41235374_41235525 | 0.26 |
ENSG00000207962 |
. |
12493 |
0.12 |
| chr6_3139599_3139904 | 0.26 |
BPHL |
biphenyl hydrolase-like (serine hydrolase) |
908 |
0.5 |
| chr5_14445022_14445319 | 0.26 |
TRIO |
trio Rho guanine nucleotide exchange factor |
43387 |
0.2 |
| chrX_153770737_153771284 | 0.26 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
532 |
0.58 |
| chr1_59602387_59602647 | 0.26 |
FGGY |
FGGY carbohydrate kinase domain containing |
159793 |
0.04 |
| chr3_185375823_185376288 | 0.25 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
44912 |
0.15 |
| chr7_73497247_73497589 | 0.25 |
LIMK1 |
LIM domain kinase 1 |
155 |
0.96 |
| chr1_61637743_61637894 | 0.25 |
RP4-802A10.1 |
|
47413 |
0.17 |
| chr17_26312086_26312243 | 0.25 |
ENSG00000251818 |
. |
33632 |
0.13 |
| chr12_48147567_48147718 | 0.25 |
SLC48A1 |
solute carrier family 48 (heme transporter), member 1 |
57 |
0.96 |
| chr4_41062724_41062875 | 0.24 |
ENSG00000199790 |
. |
23825 |
0.18 |
| chr12_15112233_15112384 | 0.24 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
1892 |
0.3 |
| chr1_95328697_95328848 | 0.24 |
SLC44A3 |
solute carrier family 44, member 3 |
4115 |
0.22 |
| chr8_97594056_97594225 | 0.24 |
SDC2 |
syndecan 2 |
1563 |
0.51 |
| chr10_26845646_26845984 | 0.24 |
ENSG00000199733 |
. |
47297 |
0.17 |
| chr2_64343006_64343324 | 0.24 |
AC074289.1 |
|
27208 |
0.19 |
| chr2_11343535_11343897 | 0.24 |
ENSG00000207267 |
. |
30398 |
0.15 |
| chr16_29280883_29281034 | 0.24 |
RP11-231C14.6 |
|
42713 |
0.13 |
| chr2_232229722_232230070 | 0.24 |
ENSG00000263641 |
. |
2477 |
0.26 |
| chr8_18953240_18953562 | 0.23 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
11161 |
0.29 |
| chr8_139855770_139855921 | 0.23 |
COL22A1 |
collagen, type XXII, alpha 1 |
64040 |
0.16 |
| chr16_73104328_73104761 | 0.23 |
ZFHX3 |
zinc finger homeobox 3 |
10947 |
0.24 |
| chr17_30822967_30823118 | 0.23 |
RP11-466A19.1 |
|
488 |
0.69 |
| chr17_62038673_62039025 | 0.23 |
SCN4A |
sodium channel, voltage-gated, type IV, alpha subunit |
11429 |
0.12 |
| chr1_32049799_32049950 | 0.23 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
7735 |
0.14 |
| chr10_114021204_114021502 | 0.23 |
TECTB |
tectorin beta |
22140 |
0.23 |
| chr3_31212208_31212359 | 0.23 |
ENSG00000265376 |
. |
9004 |
0.28 |
| chr12_52569964_52570127 | 0.23 |
KRT80 |
keratin 80 |
15739 |
0.12 |
| chr12_78450993_78451144 | 0.22 |
RP11-136F16.1 |
|
34449 |
0.22 |
| chr1_8740143_8740294 | 0.22 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
23060 |
0.23 |
| chr8_105378884_105379035 | 0.22 |
DCSTAMP |
dendrocyte expressed seven transmembrane protein |
18190 |
0.18 |
| chr22_38573907_38574208 | 0.22 |
PLA2G6 |
phospholipase A2, group VI (cytosolic, calcium-independent) |
2354 |
0.22 |
| chr17_983655_983915 | 0.22 |
ABR |
active BCR-related |
1399 |
0.45 |
| chr9_127116150_127116301 | 0.22 |
ENSG00000264237 |
. |
3415 |
0.26 |
| chr14_59721391_59721542 | 0.22 |
DAAM1 |
dishevelled associated activator of morphogenesis 1 |
8707 |
0.3 |
| chr10_50887461_50887643 | 0.22 |
C10orf53 |
chromosome 10 open reading frame 53 |
145 |
0.97 |
| chr19_6463883_6464309 | 0.22 |
CRB3 |
crumbs homolog 3 (Drosophila) |
194 |
0.84 |
| chr3_168823031_168823291 | 0.21 |
MECOM |
MDS1 and EVI1 complex locus |
22661 |
0.28 |
| chr17_40998909_40999245 | 0.21 |
AOC2 |
amine oxidase, copper containing 2 (retina-specific) |
2459 |
0.12 |
| chr6_43091623_43091783 | 0.21 |
PTK7 |
protein tyrosine kinase 7 |
6626 |
0.13 |
| chr2_174937001_174937255 | 0.21 |
SP3 |
Sp3 transcription factor |
106698 |
0.07 |
| chr11_19702330_19702481 | 0.21 |
NAV2 |
neuron navigator 2 |
32476 |
0.19 |
| chr1_183250951_183251102 | 0.21 |
NMNAT2 |
nicotinamide nucleotide adenylyltransferase 2 |
22983 |
0.23 |
| chr2_238066163_238066314 | 0.21 |
COPS8 |
COP9 signalosome subunit 8 |
71652 |
0.1 |
| chr6_15897284_15897435 | 0.21 |
MYLIP |
myosin regulatory light chain interacting protein |
231997 |
0.02 |
| chr21_43460258_43460538 | 0.21 |
UMODL1 |
uromodulin-like 1 |
22670 |
0.16 |
| chr14_33698651_33698802 | 0.21 |
NPAS3 |
neuronal PAS domain protein 3 |
14208 |
0.32 |
| chr13_33000508_33000853 | 0.21 |
N4BP2L1 |
NEDD4 binding protein 2-like 1 |
1471 |
0.43 |
| chr13_111033798_111033949 | 0.20 |
ENSG00000238629 |
. |
32679 |
0.18 |
| chr1_53601169_53601320 | 0.20 |
SLC1A7 |
solute carrier family 1 (glutamate transporter), member 7 |
7005 |
0.14 |
| chr2_102079958_102080109 | 0.20 |
RFX8 |
RFX family member 8, lacking RFX DNA binding domain |
10704 |
0.26 |
| chr8_26485560_26485711 | 0.20 |
DPYSL2 |
dihydropyrimidinase-like 2 |
49714 |
0.17 |
| chr19_47239966_47240117 | 0.20 |
ENSG00000222614 |
. |
472 |
0.64 |
| chr3_127454470_127454692 | 0.20 |
MGLL |
monoglyceride lipase |
619 |
0.78 |
| chr3_72378625_72378776 | 0.20 |
ENSG00000212070 |
. |
67121 |
0.13 |
| chr4_56042024_56042324 | 0.20 |
ENSG00000239545 |
. |
39905 |
0.15 |
| chr19_2188884_2189035 | 0.20 |
DOT1L |
DOT1-like histone H3K79 methyltransferase |
24630 |
0.08 |
| chrX_48350514_48350665 | 0.20 |
FTSJ1 |
FtsJ RNA methyltransferase homolog 1 (E. coli) |
14209 |
0.1 |
| chr16_85597214_85597712 | 0.20 |
GSE1 |
Gse1 coiled-coil protein |
47552 |
0.14 |
| chr2_120980604_120980755 | 0.20 |
TMEM185B |
transmembrane protein 185B |
305 |
0.89 |
| chr12_1579383_1579534 | 0.20 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
59599 |
0.11 |
| chr12_112043846_112043997 | 0.20 |
ATXN2 |
ataxin 2 |
6441 |
0.18 |
| chr21_39850220_39850518 | 0.20 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
19976 |
0.28 |
| chr11_62369886_62370188 | 0.20 |
MTA2 |
metastasis associated 1 family, member 2 |
725 |
0.33 |
| chr2_101507765_101507916 | 0.20 |
NPAS2 |
neuronal PAS domain protein 2 |
33768 |
0.17 |
| chr17_773553_773704 | 0.20 |
NXN |
nucleoredoxin |
6277 |
0.16 |
| chr6_157469967_157470240 | 0.19 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
58 |
0.98 |
| chr22_41050157_41050308 | 0.19 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
17537 |
0.19 |
| chr1_44705174_44705804 | 0.19 |
ERI3-IT1 |
ERI3 intronic transcript 1 (non-protein coding) |
4456 |
0.21 |
| chr2_40663401_40663552 | 0.19 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
6032 |
0.34 |
| chr7_41187429_41187580 | 0.19 |
AC005160.3 |
|
372347 |
0.01 |
| chr9_136325577_136325728 | 0.19 |
CACFD1 |
calcium channel flower domain containing 1 |
497 |
0.68 |
| chr3_127484247_127484497 | 0.19 |
MGLL |
monoglyceride lipase |
18479 |
0.22 |
| chr17_39240048_39240199 | 0.19 |
KRTAP4-7 |
keratin associated protein 4-7 |
336 |
0.68 |
| chr12_69007841_69008681 | 0.19 |
RAP1B |
RAP1B, member of RAS oncogene family |
3456 |
0.26 |
| chr21_33846975_33847126 | 0.19 |
EVA1C |
eva-1 homolog C (C. elegans) |
46408 |
0.11 |
| chr15_68645681_68645891 | 0.19 |
FEM1B |
fem-1 homolog b (C. elegans) |
63236 |
0.11 |
| chr6_43958464_43958740 | 0.19 |
C6orf223 |
chromosome 6 open reading frame 223 |
9715 |
0.17 |
| chr16_85590754_85591408 | 0.19 |
GSE1 |
Gse1 coiled-coil protein |
53934 |
0.13 |
| chr11_107799088_107799595 | 0.19 |
RAB39A |
RAB39A, member RAS oncogene family |
112 |
0.77 |
| chr15_73979466_73979617 | 0.19 |
CD276 |
CD276 molecule |
2194 |
0.36 |
| chr6_26432216_26432367 | 0.19 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
8409 |
0.11 |
| chr2_121528712_121528929 | 0.19 |
GLI2 |
GLI family zinc finger 2 |
21165 |
0.27 |
| chr7_5674087_5674709 | 0.19 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
40081 |
0.12 |
| chr13_111866660_111867080 | 0.19 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
9234 |
0.25 |
| chr17_40554278_40554429 | 0.19 |
ENSG00000221020 |
. |
3326 |
0.15 |
| chr9_36158585_36158800 | 0.18 |
CCIN |
calicin |
10697 |
0.17 |
| chr13_30567918_30568069 | 0.18 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
67205 |
0.13 |
| chr7_101524016_101524398 | 0.18 |
CTA-339C12.1 |
|
56198 |
0.12 |
| chr19_52270463_52270746 | 0.18 |
FPR2 |
formyl peptide receptor 2 |
1027 |
0.41 |
| chr8_145692200_145692386 | 0.18 |
KIFC2 |
kinesin family member C2 |
645 |
0.36 |
| chr2_121673188_121673339 | 0.18 |
GLI2 |
GLI family zinc finger 2 |
118342 |
0.06 |
| chr9_81760534_81760685 | 0.18 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
426079 |
0.01 |
| chr15_63322256_63322433 | 0.18 |
TPM1 |
tropomyosin 1 (alpha) |
12487 |
0.18 |
| chr17_7837465_7837616 | 0.18 |
CNTROB |
centrobin, centrosomal BRCA2 interacting protein |
1142 |
0.27 |
| chr10_80997764_80997915 | 0.18 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
68136 |
0.12 |
| chr10_105251971_105252122 | 0.18 |
NEURL1 |
neuralized E3 ubiquitin protein ligase 1 |
1690 |
0.23 |
| chr14_104018069_104018220 | 0.18 |
ENSG00000252469 |
. |
1462 |
0.17 |
| chr21_46852549_46852700 | 0.18 |
COL18A1-AS1 |
COL18A1 antisense RNA 1 |
7639 |
0.17 |
| chr3_8499776_8500071 | 0.18 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
43361 |
0.15 |
| chr10_81000941_81001092 | 0.18 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
64959 |
0.13 |
| chr2_30454852_30455048 | 0.18 |
LBH |
limb bud and heart development |
69 |
0.98 |
| chr21_42484961_42485351 | 0.18 |
LINC00323 |
long intergenic non-protein coding RNA 323 |
34194 |
0.15 |
| chr1_108661134_108661345 | 0.18 |
SLC25A24 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
74201 |
0.09 |
| chr11_61809719_61809884 | 0.18 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
74348 |
0.07 |
| chr20_2795832_2795989 | 0.18 |
C20orf141 |
chromosome 20 open reading frame 141 |
253 |
0.52 |
| chr12_124870235_124870636 | 0.18 |
NCOR2 |
nuclear receptor corepressor 2 |
2935 |
0.37 |
| chr13_25485405_25485556 | 0.18 |
CENPJ |
centromere protein J |
7348 |
0.24 |
| chr19_4391069_4391613 | 0.18 |
SH3GL1 |
SH3-domain GRB2-like 1 |
734 |
0.43 |
| chr1_87244168_87244361 | 0.18 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
73686 |
0.09 |
| chr8_85095605_85095756 | 0.18 |
RALYL |
RALY RNA binding protein-like |
89 |
0.99 |
| chr17_81021007_81021158 | 0.18 |
B3GNTL1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
11396 |
0.21 |
| chr2_113470510_113470661 | 0.18 |
NT5DC4 |
5'-nucleotidase domain containing 4 |
8478 |
0.2 |
| chr2_207974591_207974747 | 0.18 |
ENSG00000253008 |
. |
128 |
0.96 |
| chr22_38597727_38597878 | 0.18 |
MAFF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
87 |
0.95 |
| chr6_37961556_37961707 | 0.18 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
63896 |
0.12 |
| chr3_13549513_13549664 | 0.17 |
FBLN2 |
fibulin 2 |
24236 |
0.16 |
| chr7_92369983_92370216 | 0.17 |
ENSG00000206763 |
. |
38971 |
0.18 |
| chr13_101999259_101999410 | 0.17 |
NALCN |
sodium leak channel, non-selective |
52171 |
0.18 |
| chr3_148184411_148184664 | 0.17 |
AGTR1 |
angiotensin II receptor, type 1 |
231034 |
0.02 |
| chr9_86784994_86785145 | 0.17 |
RP11-380F14.2 |
|
108065 |
0.06 |
| chr1_23710189_23710340 | 0.17 |
TCEA3 |
transcription elongation factor A (SII), 3 |
3648 |
0.16 |
| chr3_149865017_149865176 | 0.17 |
RP11-167H9.4 |
|
49277 |
0.15 |
| chr3_8890314_8890465 | 0.17 |
RAD18 |
RAD18 homolog (S. cerevisiae) |
53693 |
0.11 |
| chr16_79204965_79205116 | 0.17 |
RP11-556H2.2 |
|
65186 |
0.11 |
| chrY_2709779_2709930 | 0.17 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
107 |
0.98 |
| chr17_39261210_39261361 | 0.17 |
KRTAP4-9 |
keratin associated protein 4-9 |
299 |
0.71 |
| chr10_135073858_135074032 | 0.17 |
MIR202HG |
MIR202 host gene (non-protein coding) |
12550 |
0.1 |
| chr14_62004306_62004579 | 0.17 |
RP11-47I22.1 |
|
7563 |
0.2 |
| chr18_494818_494969 | 0.17 |
COLEC12 |
collectin sub-family member 12 |
5829 |
0.22 |
| chr12_19699587_19699738 | 0.17 |
AEBP2 |
AE binding protein 2 |
46599 |
0.18 |
| chr8_41656318_41656469 | 0.17 |
ANK1 |
ankyrin 1, erythrocytic |
1253 |
0.45 |
| chr2_46343393_46343544 | 0.17 |
AC017006.2 |
|
37501 |
0.18 |
| chr3_46543509_46543660 | 0.17 |
RTP3 |
receptor (chemosensory) transporter protein 3 |
4603 |
0.17 |
| chr1_20820718_20821010 | 0.17 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
8151 |
0.19 |
| chr1_94127454_94127736 | 0.17 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
19331 |
0.22 |
| chr3_66083676_66083827 | 0.17 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
35534 |
0.18 |
| chr3_126398342_126398493 | 0.17 |
TXNRD3 |
thioredoxin reductase 3 |
24419 |
0.17 |
| chr1_153508734_153509122 | 0.17 |
S100A6 |
S100 calcium binding protein A6 |
208 |
0.85 |
| chr12_92378267_92378418 | 0.17 |
C12orf79 |
chromosome 12 open reading frame 79 |
152455 |
0.04 |
| chr11_116966114_116966583 | 0.17 |
SIK3 |
SIK family kinase 3 |
2639 |
0.22 |
| chr9_127051927_127052078 | 0.17 |
NEK6 |
NIMA-related kinase 6 |
2248 |
0.29 |
| chr5_145564514_145564665 | 0.16 |
LARS |
leucyl-tRNA synthetase |
2366 |
0.3 |
| chr1_113265986_113266348 | 0.16 |
FAM19A3 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A3 |
2968 |
0.15 |
| chr20_62530297_62530448 | 0.16 |
DNAJC5 |
DnaJ (Hsp40) homolog, subfamily C, member 5 |
3837 |
0.09 |
| chr12_71439781_71440156 | 0.16 |
CTD-2021H9.2 |
|
58338 |
0.13 |
| chr4_164253447_164254003 | 0.16 |
NPY1R |
neuropeptide Y receptor Y1 |
23 |
0.98 |
| chr2_201652506_201652946 | 0.16 |
ENSG00000201737 |
. |
12419 |
0.12 |
| chr22_41418613_41418788 | 0.16 |
ENSG00000222698 |
. |
22801 |
0.13 |
| chr5_41793545_41793696 | 0.16 |
OXCT1 |
3-oxoacid CoA transferase 1 |
862 |
0.71 |
| chr16_4428001_4428209 | 0.16 |
VASN |
vasorin |
6256 |
0.13 |
| chr3_25823149_25824090 | 0.16 |
NGLY1 |
N-glycanase 1 |
744 |
0.48 |
| chr6_109745594_109745745 | 0.16 |
ENSG00000239160 |
. |
9774 |
0.12 |
| chr16_57095437_57095664 | 0.16 |
RP11-322D14.2 |
|
3141 |
0.17 |
| chr18_32288595_32288746 | 0.16 |
DTNA |
dystrobrevin, alpha |
1578 |
0.51 |
| chr3_111686013_111686245 | 0.16 |
ABHD10 |
abhydrolase domain containing 10 |
11728 |
0.19 |
| chr20_43332745_43332896 | 0.16 |
RP11-445H22.3 |
|
8083 |
0.13 |
| chr5_54179191_54179535 | 0.16 |
ENSG00000221073 |
. |
30587 |
0.19 |
| chr19_15377356_15377507 | 0.16 |
BRD4 |
bromodomain containing 4 |
13831 |
0.16 |
| chr17_48718114_48718630 | 0.16 |
ABCC3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
6154 |
0.13 |
| chr1_198849285_198849436 | 0.16 |
ENSG00000207759 |
. |
21078 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 0.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.4 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.1 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.2 | GO:0061245 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 0.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.0 | GO:0060453 | regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:2000644 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.1 | GO:0001975 | response to amphetamine(GO:0001975) |
| 0.0 | 0.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0048596 | embryonic camera-type eye morphogenesis(GO:0048596) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.0 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine production(GO:0032682) negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.0 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.0 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.1 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.4 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |