Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
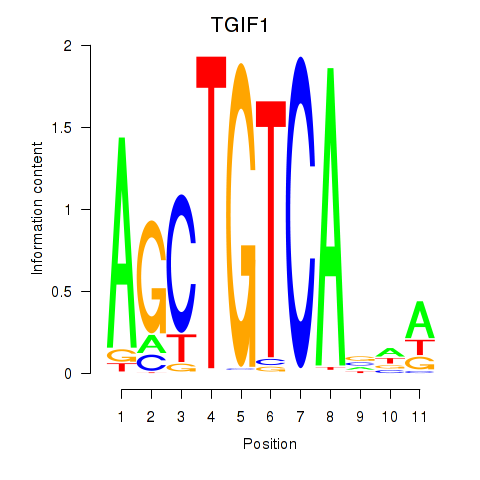
Results for TGIF1
Z-value: 0.92
Transcription factors associated with TGIF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TGIF1
|
ENSG00000177426.16 | TGFB induced factor homeobox 1 |
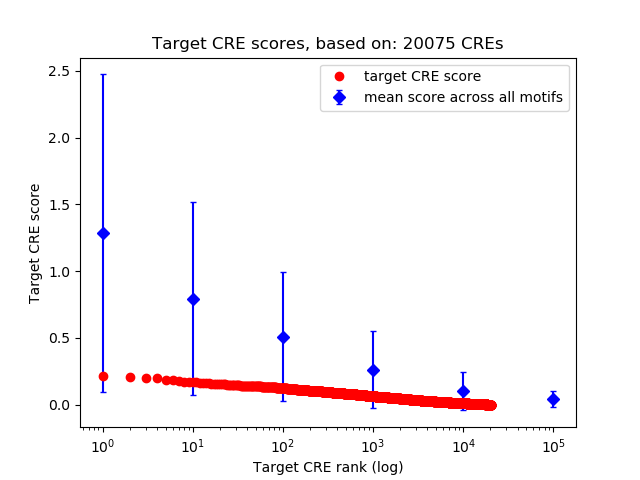
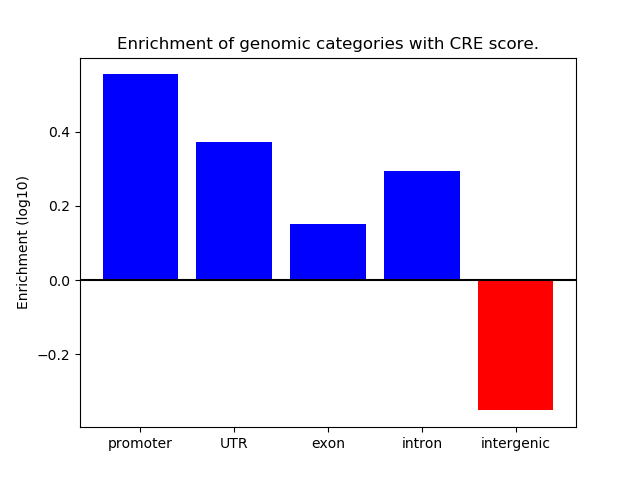
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_3340712_3340863 | TGIF1 | 70819 | 0.080968 | 0.62 | 7.6e-02 | Click! |
| chr18_3459581_3459732 | TGIF1 | 4244 | 0.247691 | -0.58 | 1.0e-01 | Click! |
| chr18_3446975_3447243 | TGIF1 | 498 | 0.819288 | 0.54 | 1.3e-01 | Click! |
| chr18_3478848_3478999 | TGIF1 | 23511 | 0.181872 | -0.53 | 1.4e-01 | Click! |
| chr18_3448954_3449105 | TGIF1 | 382 | 0.877412 | 0.52 | 1.5e-01 | Click! |
Activity of the TGIF1 motif across conditions
Conditions sorted by the z-value of the TGIF1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
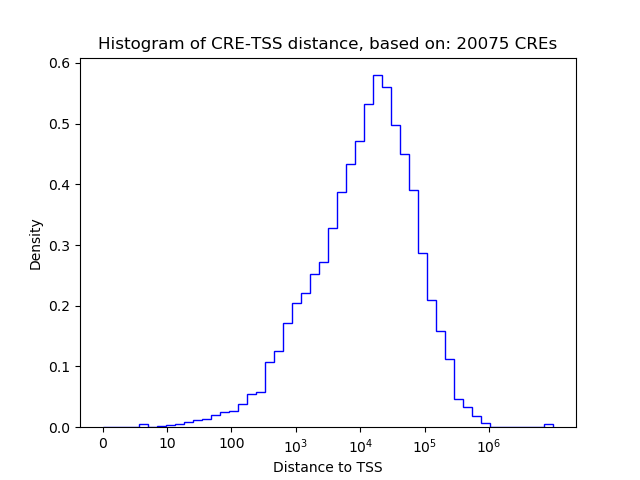
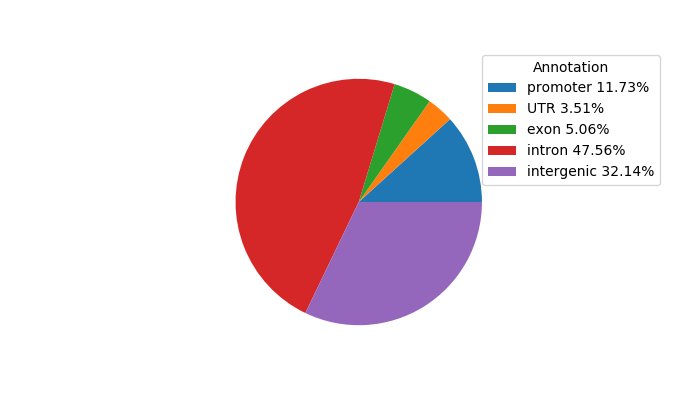
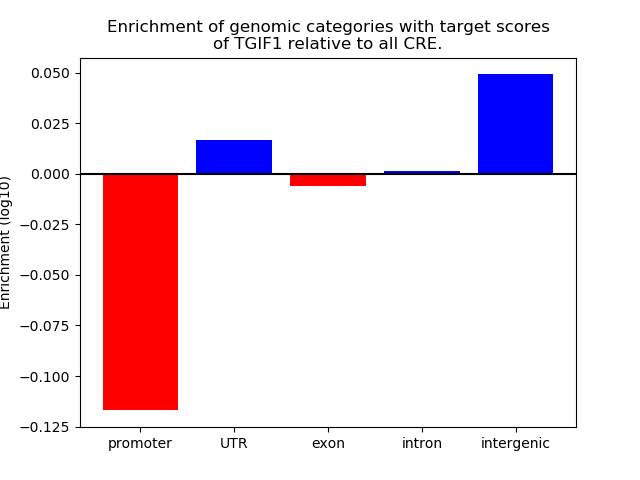
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_33208774_33208970 | 0.21 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
11441 |
0.29 |
| chr22_36000465_36000757 | 0.21 |
MB |
myoglobin |
11149 |
0.19 |
| chr1_156196195_156196346 | 0.20 |
PMF1-BGLAP |
PMF1-BGLAP readthrough |
13470 |
0.09 |
| chr5_167532981_167533278 | 0.20 |
ENSG00000253065 |
. |
72919 |
0.1 |
| chr21_34461417_34461568 | 0.19 |
AP000282.2 |
|
18082 |
0.16 |
| chr7_4166622_4166773 | 0.18 |
SDK1 |
sidekick cell adhesion molecule 1 |
176145 |
0.03 |
| chr1_156509588_156509739 | 0.18 |
ENSG00000238843 |
. |
10541 |
0.11 |
| chr10_34004252_34004403 | 0.17 |
NRP1 |
neuropilin 1 |
379137 |
0.01 |
| chr4_48900915_48901066 | 0.17 |
OCIAD2 |
OCIA domain containing 2 |
7784 |
0.2 |
| chr21_41842867_41843018 | 0.17 |
ENSG00000207147 |
. |
42129 |
0.18 |
| chr7_10018425_10018576 | 0.17 |
ENSG00000212422 |
. |
243924 |
0.02 |
| chr9_90146510_90146661 | 0.17 |
DAPK1-IT1 |
DAPK1 intronic transcript 1 (non-protein coding) |
21784 |
0.22 |
| chr8_19243061_19243460 | 0.16 |
SH2D4A |
SH2 domain containing 4A |
66159 |
0.14 |
| chr11_12194861_12195012 | 0.16 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
11233 |
0.26 |
| chr1_185135340_185135491 | 0.16 |
SWT1 |
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
8801 |
0.19 |
| chr13_92002817_92003060 | 0.16 |
ENSG00000215417 |
. |
59 |
0.99 |
| chr16_49534864_49535143 | 0.16 |
C16orf78 |
chromosome 16 open reading frame 78 |
127269 |
0.05 |
| chr2_45735351_45735502 | 0.16 |
SRBD1 |
S1 RNA binding domain 1 |
59728 |
0.13 |
| chr20_57601860_57602011 | 0.16 |
ATP5E |
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
5412 |
0.15 |
| chr8_93693450_93693601 | 0.15 |
ENSG00000221172 |
. |
45962 |
0.18 |
| chr2_207380556_207380707 | 0.15 |
ADAM23 |
ADAM metallopeptidase domain 23 |
72040 |
0.1 |
| chr5_56784424_56784602 | 0.15 |
ACTBL2 |
actin, beta-like 2 |
5877 |
0.24 |
| chr10_50390146_50390361 | 0.15 |
C10orf128 |
chromosome 10 open reading frame 128 |
6104 |
0.2 |
| chr16_68381624_68381775 | 0.15 |
ENSG00000222177 |
. |
18267 |
0.09 |
| chr4_6013606_6013757 | 0.15 |
C4orf50 |
chromosome 4 open reading frame 50 |
22136 |
0.24 |
| chr3_168334920_168335071 | 0.15 |
ENSG00000207717 |
. |
65353 |
0.15 |
| chr15_41925877_41926028 | 0.15 |
MGA |
MGA, MAX dimerization protein |
12218 |
0.18 |
| chr8_121016842_121017083 | 0.15 |
COL14A1 |
collagen, type XIV, alpha 1 |
100854 |
0.07 |
| chr7_40796116_40796267 | 0.15 |
AC005160.3 |
|
18966 |
0.3 |
| chr15_68634684_68634835 | 0.15 |
FEM1B |
fem-1 homolog b (C. elegans) |
52209 |
0.13 |
| chr9_113403073_113403248 | 0.15 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
27891 |
0.21 |
| chr11_44938433_44938696 | 0.15 |
TSPAN18 |
tetraspanin 18 |
10606 |
0.25 |
| chr3_16513920_16514071 | 0.14 |
RFTN1 |
raftlin, lipid raft linker 1 |
10377 |
0.26 |
| chr6_20564770_20564937 | 0.14 |
CDKAL1 |
CDK5 regulatory subunit associated protein 1-like 1 |
18281 |
0.22 |
| chr11_20029031_20029182 | 0.14 |
NAV2 |
neuron navigator 2 |
14996 |
0.2 |
| chr1_83171006_83171157 | 0.14 |
LPHN2 |
latrophilin 2 |
725508 |
0.0 |
| chr11_62321469_62322295 | 0.14 |
AHNAK |
AHNAK nucleoprotein |
1825 |
0.17 |
| chr6_109162296_109162639 | 0.14 |
ARMC2 |
armadillo repeat containing 2 |
7152 |
0.28 |
| chr1_145759757_145760123 | 0.14 |
PDZK1 |
PDZ domain containing 1 |
16677 |
0.17 |
| chr20_39561263_39561414 | 0.14 |
ENSG00000238908 |
. |
78469 |
0.1 |
| chr13_21212008_21212159 | 0.14 |
ENSG00000222726 |
. |
25783 |
0.18 |
| chr8_116661022_116661259 | 0.14 |
TRPS1 |
trichorhinophalangeal syndrome I |
12765 |
0.31 |
| chr3_121590668_121590819 | 0.14 |
SLC15A2 |
solute carrier family 15 (oligopeptide transporter), member 2 |
22193 |
0.15 |
| chr6_161454710_161454940 | 0.14 |
MAP3K4 |
mitogen-activated protein kinase kinase kinase 4 |
11599 |
0.25 |
| chr6_82193288_82193439 | 0.14 |
RP1-300G12.2 |
|
51267 |
0.16 |
| chr11_95452659_95452810 | 0.14 |
FAM76B |
family with sequence similarity 76, member B |
67235 |
0.12 |
| chr1_157717360_157717511 | 0.14 |
FCRL2 |
Fc receptor-like 2 |
20129 |
0.18 |
| chr12_124230277_124230428 | 0.14 |
ATP6V0A2 |
ATPase, H+ transporting, lysosomal V0 subunit a2 |
4191 |
0.2 |
| chr9_12697396_12697547 | 0.14 |
TYRP1 |
tyrosinase-related protein 1 |
1751 |
0.46 |
| chr7_150360680_150361171 | 0.14 |
GIMAP2 |
GTPase, IMAP family member 2 |
21863 |
0.15 |
| chr13_103935436_103935587 | 0.14 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
216315 |
0.02 |
| chr21_34426463_34426615 | 0.14 |
OLIG1 |
oligodendrocyte transcription factor 1 |
15911 |
0.16 |
| chr1_245084915_245085066 | 0.14 |
ENSG00000201758 |
. |
22222 |
0.14 |
| chr15_82235742_82235893 | 0.14 |
ENSG00000222521 |
. |
100818 |
0.07 |
| chr1_239701671_239701822 | 0.14 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
90627 |
0.1 |
| chr21_26946893_26947044 | 0.14 |
ENSG00000234883 |
. |
676 |
0.69 |
| chr4_159703546_159703697 | 0.14 |
ENSG00000206703 |
. |
12227 |
0.18 |
| chr16_79449443_79449594 | 0.14 |
ENSG00000222244 |
. |
151167 |
0.04 |
| chr14_70615527_70615823 | 0.14 |
SLC8A3 |
solute carrier family 8 (sodium/calcium exchanger), member 3 |
19526 |
0.23 |
| chr1_242012350_242012723 | 0.14 |
EXO1 |
exonuclease 1 |
283 |
0.94 |
| chr1_66556636_66556787 | 0.14 |
ENSG00000223152 |
. |
3518 |
0.3 |
| chr1_168803043_168803442 | 0.14 |
ENSG00000221578 |
. |
86122 |
0.09 |
| chr8_68632907_68633058 | 0.14 |
ENSG00000221660 |
. |
9115 |
0.21 |
| chr2_176417653_176417879 | 0.14 |
ENSG00000221347 |
. |
222665 |
0.02 |
| chr3_194838476_194838628 | 0.14 |
XXYLT1 |
xyloside xylosyltransferase 1 |
4369 |
0.17 |
| chr18_8455726_8455877 | 0.14 |
ENSG00000242985 |
. |
16028 |
0.18 |
| chr17_27419224_27419375 | 0.14 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
762 |
0.58 |
| chr19_16228666_16229162 | 0.13 |
RAB8A |
RAB8A, member RAS oncogene family |
6207 |
0.14 |
| chr9_80517904_80518204 | 0.13 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
80139 |
0.11 |
| chr6_6656190_6656366 | 0.13 |
LY86-AS1 |
LY86 antisense RNA 1 |
33274 |
0.22 |
| chr12_9921460_9921827 | 0.13 |
CD69 |
CD69 molecule |
8146 |
0.16 |
| chr12_49213958_49214109 | 0.13 |
CACNB3 |
calcium channel, voltage-dependent, beta 3 subunit |
1341 |
0.26 |
| chr4_116213074_116213225 | 0.13 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
178117 |
0.03 |
| chr2_165752418_165752569 | 0.13 |
ENSG00000223318 |
. |
206 |
0.94 |
| chr16_23908795_23909009 | 0.13 |
PRKCB |
protein kinase C, beta |
60358 |
0.13 |
| chr6_13002773_13002924 | 0.13 |
PHACTR1 |
phosphatase and actin regulator 1 |
44634 |
0.2 |
| chr2_130569309_130569460 | 0.13 |
AC079776.2 |
|
113914 |
0.06 |
| chr12_68285414_68285565 | 0.13 |
IFNG-AS1 |
IFNG antisense RNA 1 |
97736 |
0.09 |
| chr15_38988151_38988528 | 0.13 |
C15orf53 |
chromosome 15 open reading frame 53 |
460 |
0.89 |
| chr18_53903146_53903297 | 0.13 |
ENSG00000201816 |
. |
156396 |
0.04 |
| chr10_112478002_112478221 | 0.13 |
ENSG00000252036 |
. |
38103 |
0.13 |
| chr17_49043165_49043316 | 0.13 |
SPAG9 |
sperm associated antigen 9 |
28058 |
0.17 |
| chr6_109660786_109660937 | 0.13 |
ENSG00000201023 |
. |
34164 |
0.12 |
| chr6_45789023_45789174 | 0.13 |
ENSG00000252738 |
. |
175257 |
0.03 |
| chr14_72595830_72595981 | 0.13 |
ENSG00000200298 |
. |
106672 |
0.07 |
| chr7_101539600_101539793 | 0.13 |
CTB-181H17.1 |
|
63700 |
0.11 |
| chr11_42274588_42274739 | 0.13 |
LRRC4C |
leucine rich repeat containing 4C |
793340 |
0.0 |
| chr17_78845626_78846398 | 0.13 |
RPTOR |
regulatory associated protein of MTOR, complex 1 |
50512 |
0.1 |
| chr14_49949021_49949172 | 0.13 |
ENSG00000252424 |
. |
70255 |
0.08 |
| chr6_44796997_44797148 | 0.13 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
126175 |
0.06 |
| chr17_1870594_1870879 | 0.13 |
CTD-2545H1.2 |
|
28172 |
0.1 |
| chr10_8374212_8374363 | 0.13 |
GATA3 |
GATA binding protein 3 |
277518 |
0.01 |
| chr3_138607537_138607828 | 0.13 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
53902 |
0.11 |
| chr3_179415944_179416095 | 0.13 |
USP13 |
ubiquitin specific peptidase 13 (isopeptidase T-3) |
23333 |
0.19 |
| chr12_125045629_125045780 | 0.13 |
NCOR2 |
nuclear receptor corepressor 2 |
6306 |
0.32 |
| chr2_165650853_165651004 | 0.13 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
20664 |
0.19 |
| chr18_10899035_10899186 | 0.13 |
RP11-513M1.1 |
|
5495 |
0.29 |
| chr2_153446163_153446314 | 0.12 |
FMNL2 |
formin-like 2 |
29854 |
0.24 |
| chr5_148153495_148153674 | 0.12 |
ADRB2 |
adrenoceptor beta 2, surface |
52572 |
0.15 |
| chrX_77829230_77829561 | 0.12 |
ZCCHC5 |
zinc finger, CCHC domain containing 5 |
85430 |
0.1 |
| chr9_80520526_80520677 | 0.12 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
82686 |
0.11 |
| chr17_55510110_55510261 | 0.12 |
ENSG00000263902 |
. |
20623 |
0.23 |
| chr5_88113170_88113321 | 0.12 |
MEF2C |
myocyte enhancer factor 2C |
6360 |
0.28 |
| chr12_4386840_4387020 | 0.12 |
CCND2-AS1 |
CCND2 antisense RNA 1 |
1580 |
0.28 |
| chr13_30147951_30148102 | 0.12 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
12903 |
0.28 |
| chr8_125648788_125648939 | 0.12 |
RP11-532M24.1 |
|
56991 |
0.12 |
| chr15_38945205_38945356 | 0.12 |
C15orf53 |
chromosome 15 open reading frame 53 |
43519 |
0.17 |
| chr8_103092530_103092681 | 0.12 |
NCALD |
neurocalcin delta |
43573 |
0.16 |
| chr1_192796399_192796550 | 0.12 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
18303 |
0.23 |
| chrX_48657873_48658024 | 0.12 |
HDAC6 |
histone deacetylase 6 |
1836 |
0.2 |
| chrX_15769905_15770141 | 0.12 |
CA5B |
carbonic anhydrase VB, mitochondrial |
1930 |
0.32 |
| chr17_1087827_1087978 | 0.12 |
ABR |
active BCR-related |
2714 |
0.24 |
| chr2_135438010_135438325 | 0.12 |
TMEM163 |
transmembrane protein 163 |
38403 |
0.21 |
| chr4_146542708_146542859 | 0.12 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
3368 |
0.26 |
| chr22_47067865_47068016 | 0.12 |
GRAMD4 |
GRAM domain containing 4 |
2565 |
0.33 |
| chr14_104321166_104321317 | 0.12 |
PPP1R13B |
protein phosphatase 1, regulatory subunit 13B |
7314 |
0.17 |
| chr3_32864793_32864944 | 0.12 |
TRIM71 |
tripartite motif containing 71, E3 ubiquitin protein ligase |
5358 |
0.28 |
| chr11_129201902_129202053 | 0.12 |
BARX2 |
BARX homeobox 2 |
43858 |
0.17 |
| chr6_13529066_13529217 | 0.12 |
ENSG00000202351 |
. |
18795 |
0.14 |
| chr1_92812169_92812320 | 0.12 |
GLMN |
glomulin, FKBP associated protein |
47700 |
0.12 |
| chr10_18937367_18937518 | 0.12 |
NSUN6 |
NOP2/Sun domain family, member 6 |
3109 |
0.24 |
| chr2_182062690_182062873 | 0.12 |
ENSG00000266705 |
. |
107598 |
0.07 |
| chr13_59042867_59043018 | 0.12 |
ENSG00000222733 |
. |
58943 |
0.17 |
| chr14_59423768_59423919 | 0.12 |
ENSG00000221427 |
. |
185879 |
0.03 |
| chr13_91864910_91865061 | 0.12 |
ENSG00000215417 |
. |
137874 |
0.05 |
| chr6_41301582_41301834 | 0.12 |
NCR2 |
natural cytotoxicity triggering receptor 2 |
1685 |
0.32 |
| chr7_136611451_136611602 | 0.12 |
hsa-mir-490 |
hsa-mir-490 |
12477 |
0.21 |
| chr1_15500617_15500768 | 0.12 |
C1orf195 |
chromosome 1 open reading frame 195 |
2879 |
0.29 |
| chr4_79703843_79703994 | 0.12 |
BMP2K |
BMP2 inducible kinase |
6386 |
0.2 |
| chr20_11229977_11230245 | 0.12 |
C20orf187 |
chromosome 20 open reading frame 187 |
221300 |
0.02 |
| chr2_65132087_65132267 | 0.12 |
ENSG00000244534 |
. |
2095 |
0.28 |
| chr20_1008398_1008549 | 0.12 |
RSPO4 |
R-spondin 4 |
25566 |
0.2 |
| chr2_62222707_62222858 | 0.12 |
COMMD1 |
copper metabolism (Murr1) domain containing 1 |
5332 |
0.29 |
| chr19_33236480_33236702 | 0.12 |
TDRD12 |
tudor domain containing 12 |
25612 |
0.14 |
| chr3_193952906_193953057 | 0.12 |
ENSG00000242201 |
. |
89518 |
0.07 |
| chr5_31931833_31931984 | 0.12 |
ENSG00000266243 |
. |
4357 |
0.25 |
| chr1_110703215_110703366 | 0.12 |
RP5-1028L10.1 |
|
6515 |
0.13 |
| chr9_131832684_131832835 | 0.12 |
DOLPP1 |
dolichyldiphosphatase 1 |
10620 |
0.11 |
| chr7_50363677_50363924 | 0.12 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
3445 |
0.35 |
| chr17_55457411_55457562 | 0.11 |
ENSG00000263902 |
. |
32076 |
0.2 |
| chr4_88966383_88966534 | 0.11 |
PKD2 |
polycystic kidney disease 2 (autosomal dominant) |
2045 |
0.35 |
| chr3_58886323_58886474 | 0.11 |
C3orf67 |
chromosome 3 open reading frame 67 |
37090 |
0.2 |
| chr8_50160983_50161134 | 0.11 |
ENSG00000199640 |
. |
81711 |
0.11 |
| chr7_105495026_105495183 | 0.11 |
ATXN7L1 |
ataxin 7-like 1 |
21819 |
0.26 |
| chr1_37195687_37195944 | 0.11 |
RP4-614N24.1 |
|
42926 |
0.19 |
| chr7_114615058_114615617 | 0.11 |
MDFIC |
MyoD family inhibitor domain containing |
41413 |
0.22 |
| chr3_68139067_68139395 | 0.11 |
FAM19A1 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 |
83858 |
0.11 |
| chr12_98812168_98812319 | 0.11 |
ENSG00000201296 |
. |
427 |
0.88 |
| chr5_56797760_56797911 | 0.11 |
ACTBL2 |
actin, beta-like 2 |
19199 |
0.21 |
| chr7_150542662_150542813 | 0.11 |
AOC1 |
amine oxidase, copper containing 1 |
6360 |
0.19 |
| chr5_10594062_10594213 | 0.11 |
ANKRD33B |
ankyrin repeat domain 33B |
29557 |
0.17 |
| chr1_204430993_204431895 | 0.11 |
PIK3C2B |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
5030 |
0.21 |
| chr5_58448897_58449048 | 0.11 |
RP11-266N13.2 |
|
113384 |
0.07 |
| chr5_78088517_78088668 | 0.11 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
143944 |
0.05 |
| chrX_135829438_135829709 | 0.11 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
19929 |
0.17 |
| chr2_162815067_162815218 | 0.11 |
ENSG00000253046 |
. |
39850 |
0.19 |
| chr2_192696442_192696593 | 0.11 |
AC098617.1 |
|
14748 |
0.2 |
| chr15_60359286_60359437 | 0.11 |
FOXB1 |
forkhead box B1 |
62940 |
0.15 |
| chr12_39267342_39267493 | 0.11 |
CPNE8 |
copine VIII |
32016 |
0.17 |
| chr6_27235927_27236078 | 0.11 |
PRSS16 |
protease, serine, 16 (thymus) |
19287 |
0.18 |
| chr4_22905571_22905722 | 0.11 |
GPR125 |
G protein-coupled receptor 125 |
387969 |
0.01 |
| chr9_113681977_113682128 | 0.11 |
ENSG00000207401 |
. |
14634 |
0.21 |
| chr19_5119356_5119507 | 0.11 |
KDM4B |
lysine (K)-specific demethylase 4B |
36968 |
0.16 |
| chr7_142425935_142426150 | 0.11 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
31277 |
0.16 |
| chr10_71224348_71224499 | 0.11 |
TSPAN15 |
tetraspanin 15 |
13194 |
0.21 |
| chr20_37433246_37433545 | 0.11 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
953 |
0.57 |
| chr17_16939284_16939435 | 0.11 |
MPRIP |
myosin phosphatase Rho interacting protein |
6500 |
0.2 |
| chr7_20278384_20278535 | 0.11 |
MACC1 |
metastasis associated in colon cancer 1 |
21432 |
0.22 |
| chr1_16024310_16024604 | 0.11 |
ENSG00000264048 |
. |
13490 |
0.1 |
| chr7_139250491_139250642 | 0.11 |
CLEC2L |
C-type lectin domain family 2, member L |
41738 |
0.17 |
| chr15_38987903_38988075 | 0.11 |
C15orf53 |
chromosome 15 open reading frame 53 |
810 |
0.76 |
| chr9_139555674_139555825 | 0.11 |
RP11-251M1.1 |
|
876 |
0.35 |
| chr4_40154248_40154664 | 0.11 |
RP11-395I6.3 |
|
13839 |
0.18 |
| chr3_155364086_155364237 | 0.11 |
PLCH1 |
phospholipase C, eta 1 |
29667 |
0.22 |
| chr11_43399109_43399260 | 0.11 |
RP11-484D2.5 |
|
8037 |
0.16 |
| chr4_56753882_56754183 | 0.11 |
ENSG00000202358 |
. |
2271 |
0.3 |
| chr5_61627825_61628088 | 0.11 |
KIF2A |
kinesin heavy chain member 2A |
20316 |
0.22 |
| chr11_117816071_117816391 | 0.11 |
TMPRSS13 |
transmembrane protease, serine 13 |
16057 |
0.16 |
| chr17_61505350_61505582 | 0.11 |
RP11-269G24.3 |
|
2984 |
0.18 |
| chr6_49630348_49630499 | 0.11 |
RHAG |
Rh-associated glycoprotein |
25871 |
0.18 |
| chr2_23726807_23726958 | 0.11 |
AC011239.1 |
Uncharacterized protein |
20332 |
0.27 |
| chr2_60165263_60165480 | 0.11 |
ENSG00000252726 |
. |
243474 |
0.02 |
| chr4_109280961_109281363 | 0.11 |
ENSG00000232021 |
. |
183718 |
0.03 |
| chr12_27610504_27610655 | 0.11 |
SMCO2 |
single-pass membrane protein with coiled-coil domains 2 |
9164 |
0.2 |
| chr11_110341183_110341334 | 0.11 |
FDX1 |
ferredoxin 1 |
40651 |
0.21 |
| chr10_104373497_104373648 | 0.11 |
ENSG00000207029 |
. |
22291 |
0.14 |
| chr9_90954471_90954622 | 0.11 |
ENSG00000252299 |
. |
34638 |
0.2 |
| chr2_129394485_129394636 | 0.11 |
ENSG00000238379 |
. |
191788 |
0.03 |
| chr9_126298972_126299123 | 0.11 |
RP11-230L22.4 |
|
44884 |
0.15 |
| chr15_78710532_78710683 | 0.11 |
IREB2 |
iron-responsive element binding protein 2 |
19166 |
0.16 |
| chr21_40351476_40351627 | 0.11 |
ENSG00000272015 |
. |
84842 |
0.09 |
| chr13_27559343_27559515 | 0.11 |
USP12-AS1 |
USP12 antisense RNA 1 |
177563 |
0.03 |
| chr19_18140574_18140725 | 0.11 |
ARRDC2 |
arrestin domain containing 2 |
21672 |
0.13 |
| chr7_24931121_24931272 | 0.11 |
OSBPL3 |
oxysterol binding protein-like 3 |
998 |
0.62 |
| chr12_27171161_27171358 | 0.11 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
3892 |
0.18 |
| chr8_20880900_20881051 | 0.11 |
ENSG00000215945 |
. |
132667 |
0.06 |
| chr17_80436915_80437272 | 0.11 |
RP13-991F5.2 |
|
1293 |
0.23 |
| chr7_137683644_137683795 | 0.11 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
3074 |
0.26 |
| chrX_109389946_109390097 | 0.11 |
ENSG00000265584 |
. |
64675 |
0.11 |
| chr3_111021000_111021297 | 0.11 |
CD96 |
CD96 molecule |
9582 |
0.3 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.0 | GO:0043301 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.1 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0003010 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:1904063 | negative regulation of cation transmembrane transport(GO:1904063) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0042597 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |