Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
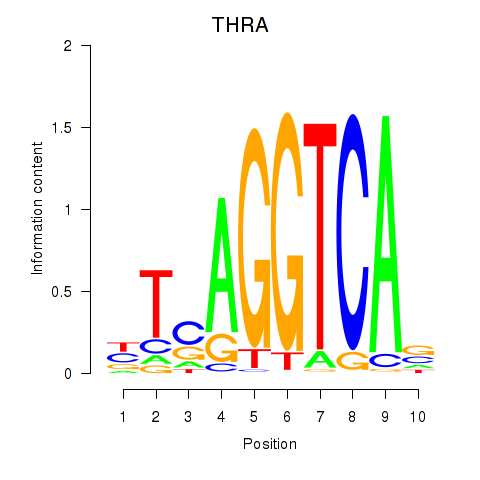
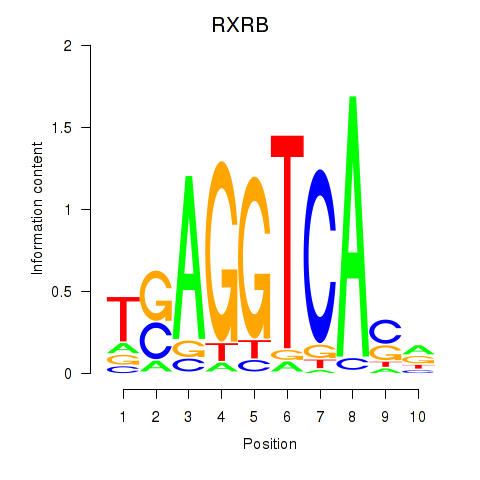
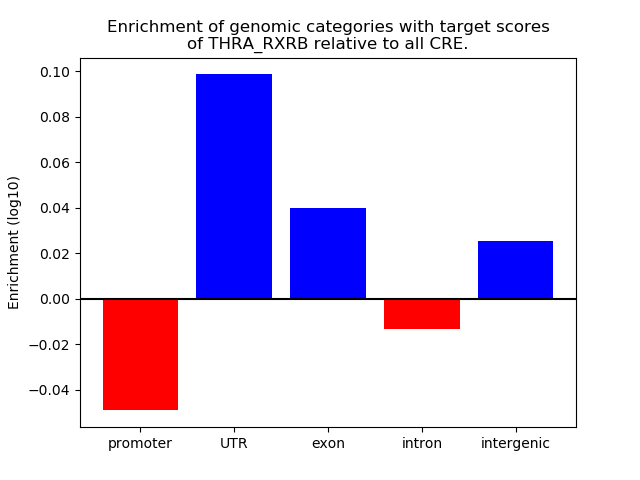
Results for THRA_RXRB
Z-value: 1.35
Transcription factors associated with THRA_RXRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRA
|
ENSG00000126351.8 | thyroid hormone receptor alpha |
|
RXRB
|
ENSG00000204231.6 | retinoid X receptor beta |
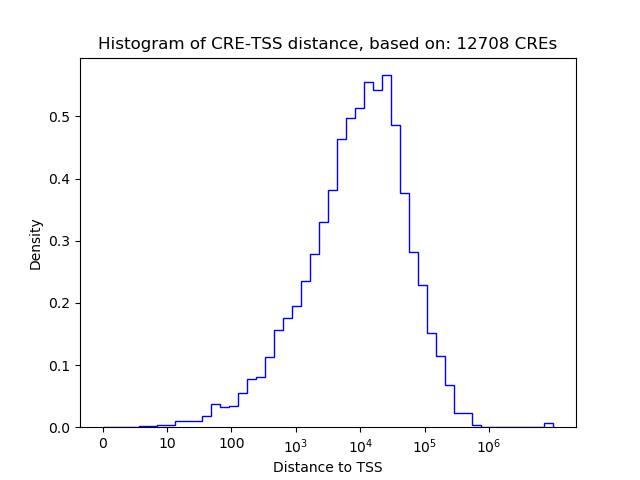
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_38218268_38218459 | THRA | 83 | 0.920685 | 0.71 | 3.3e-02 | Click! |
| chr17_38229780_38229931 | THRA | 1511 | 0.274048 | 0.70 | 3.5e-02 | Click! |
| chr17_38218492_38218643 | THRA | 121 | 0.914171 | 0.60 | 8.7e-02 | Click! |
| chr17_38232565_38232716 | THRA | 4296 | 0.136826 | 0.57 | 1.1e-01 | Click! |
| chr17_38223510_38223773 | THRA | 1515 | 0.271184 | 0.51 | 1.6e-01 | Click! |
Activity of the THRA_RXRB motif across conditions
Conditions sorted by the z-value of the THRA_RXRB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
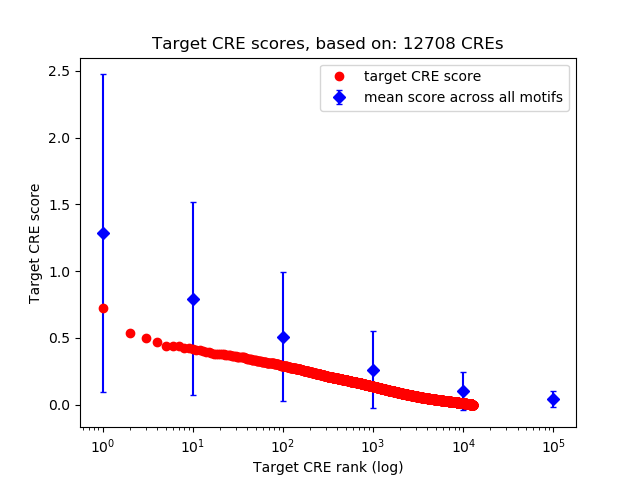
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_107819281_107819678 | 0.72 |
CD47 |
CD47 molecule |
9607 |
0.3 |
| chr2_127422232_127422532 | 0.53 |
GYPC |
glycophorin C (Gerbich blood group) |
8622 |
0.27 |
| chr15_78336776_78336956 | 0.50 |
ENSG00000221476 |
. |
5993 |
0.15 |
| chr7_150453228_150453688 | 0.47 |
GIMAP5 |
GTPase, IMAP family member 5 |
19022 |
0.14 |
| chr6_36647112_36647373 | 0.44 |
CDKN1A |
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
744 |
0.58 |
| chr3_13023386_13023537 | 0.44 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
5075 |
0.29 |
| chr10_129678837_129678988 | 0.44 |
CLRN3 |
clarin 3 |
12299 |
0.22 |
| chr2_7050734_7050964 | 0.43 |
RNF144A |
ring finger protein 144A |
6674 |
0.18 |
| chr15_85264382_85264668 | 0.42 |
SEC11A |
SEC11 homolog A (S. cerevisiae) |
4578 |
0.15 |
| chr11_118099433_118099584 | 0.42 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
3699 |
0.17 |
| chrX_48543101_48543693 | 0.41 |
WAS |
Wiskott-Aldrich syndrome |
1229 |
0.35 |
| chr20_2722949_2723333 | 0.41 |
EBF4 |
early B-cell factor 4 |
36254 |
0.09 |
| chr17_66344922_66345112 | 0.41 |
ARSG |
arylsulfatase G |
57358 |
0.1 |
| chr1_200854769_200854937 | 0.39 |
C1orf106 |
chromosome 1 open reading frame 106 |
5323 |
0.19 |
| chr6_116785564_116786071 | 0.39 |
RP1-93H18.6 |
|
962 |
0.41 |
| chr17_8859556_8859835 | 0.39 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
9329 |
0.24 |
| chr17_75913110_75913444 | 0.38 |
ENSG00000238898 |
. |
2903 |
0.3 |
| chr5_126308801_126309244 | 0.38 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
55102 |
0.15 |
| chr12_124491346_124491497 | 0.38 |
ZNF664 |
zinc finger protein 664 |
33579 |
0.08 |
| chr12_93981713_93981864 | 0.38 |
SOCS2 |
suppressor of cytokine signaling 2 |
12954 |
0.18 |
| chr5_126312253_126312404 | 0.38 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
54172 |
0.15 |
| chr17_75840839_75841130 | 0.38 |
FLJ45079 |
|
37675 |
0.18 |
| chr21_44010697_44010940 | 0.38 |
AP001625.6 |
|
28774 |
0.14 |
| chr1_155916745_155916975 | 0.37 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
4679 |
0.1 |
| chr2_174328085_174328236 | 0.37 |
CDCA7 |
cell division cycle associated 7 |
108560 |
0.07 |
| chr3_107791032_107791183 | 0.37 |
CD47 |
CD47 molecule |
12728 |
0.3 |
| chr4_38916757_38916925 | 0.37 |
ENSG00000207944 |
. |
47188 |
0.11 |
| chr6_159461814_159462217 | 0.37 |
TAGAP |
T-cell activation RhoGTPase activating protein |
4035 |
0.22 |
| chr8_144939517_144939748 | 0.36 |
EPPK1 |
epiplakin 1 |
13000 |
0.09 |
| chr16_84641040_84641383 | 0.36 |
COTL1 |
coactosin-like 1 (Dictyostelium) |
10454 |
0.17 |
| chr15_70744544_70744849 | 0.36 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
249924 |
0.02 |
| chr2_10518114_10518365 | 0.36 |
HPCAL1 |
hippocalcin-like 1 |
41908 |
0.14 |
| chr16_30468718_30468869 | 0.36 |
ENSG00000202476 |
. |
3375 |
0.08 |
| chr8_42000299_42000484 | 0.36 |
RP11-589C21.5 |
|
9890 |
0.17 |
| chr3_150888432_150888616 | 0.36 |
ENSG00000199994 |
. |
17362 |
0.16 |
| chr5_176783043_176783194 | 0.36 |
RGS14 |
regulator of G-protein signaling 14 |
1720 |
0.2 |
| chrX_128921545_128921805 | 0.35 |
SASH3 |
SAM and SH3 domain containing 3 |
7715 |
0.2 |
| chr1_1830831_1831010 | 0.35 |
RP1-140A9.1 |
|
8010 |
0.13 |
| chr9_86908771_86909182 | 0.35 |
RP11-380F14.2 |
|
15842 |
0.25 |
| chr2_99087636_99087787 | 0.35 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
26298 |
0.2 |
| chr2_198072900_198073187 | 0.34 |
ANKRD44 |
ankyrin repeat domain 44 |
10281 |
0.21 |
| chr16_29672717_29672868 | 0.34 |
SPN |
sialophorin |
1508 |
0.26 |
| chr14_90148740_90149294 | 0.34 |
ENSG00000200312 |
. |
29838 |
0.14 |
| chr1_169574716_169574995 | 0.34 |
SELP |
selectin P (granule membrane protein 140kDa, antigen CD62) |
13602 |
0.2 |
| chr5_132483935_132484150 | 0.34 |
CTB-49A3.2 |
|
12088 |
0.22 |
| chr15_76222456_76222700 | 0.34 |
FBXO22 |
F-box protein 22 |
2563 |
0.27 |
| chr8_135324203_135324532 | 0.34 |
ZFAT |
zinc finger and AT hook domain containing |
198058 |
0.03 |
| chr13_100071950_100072184 | 0.33 |
ENSG00000266207 |
. |
33656 |
0.15 |
| chr1_109188369_109188646 | 0.33 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
15170 |
0.19 |
| chr7_138731189_138731354 | 0.33 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
10496 |
0.21 |
| chr3_39300527_39300690 | 0.33 |
CX3CR1 |
chemokine (C-X3-C motif) receptor 1 |
20917 |
0.18 |
| chr7_150359146_150359387 | 0.33 |
GIMAP2 |
GTPase, IMAP family member 2 |
23522 |
0.15 |
| chr14_100546662_100546850 | 0.33 |
CTD-2376I20.1 |
|
5541 |
0.16 |
| chr14_104855874_104856139 | 0.33 |
ENSG00000222761 |
. |
11379 |
0.26 |
| chr5_133467618_133467806 | 0.33 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
8403 |
0.21 |
| chr17_56247502_56247750 | 0.33 |
OR4D2 |
olfactory receptor, family 4, subfamily D, member 2 |
609 |
0.66 |
| chr1_9717399_9717821 | 0.33 |
C1orf200 |
chromosome 1 open reading frame 200 |
2966 |
0.22 |
| chr6_90793986_90794257 | 0.32 |
ENSG00000222078 |
. |
82896 |
0.09 |
| chr1_236101876_236102050 | 0.32 |
ENSG00000206803 |
. |
23248 |
0.18 |
| chr15_91372602_91372985 | 0.32 |
CTD-3094K11.1 |
|
10161 |
0.13 |
| chr9_135992327_135992532 | 0.32 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
4134 |
0.16 |
| chr7_37393575_37393854 | 0.32 |
ELMO1 |
engulfment and cell motility 1 |
442 |
0.83 |
| chr11_67414919_67415070 | 0.32 |
ACY3 |
aspartoacylase (aminocyclase) 3 |
60 |
0.95 |
| chr11_63973377_63973615 | 0.32 |
FERMT3 |
fermitin family member 3 |
654 |
0.44 |
| chr15_64750062_64750402 | 0.32 |
ZNF609 |
zinc finger protein 609 |
2709 |
0.18 |
| chr2_135433313_135433464 | 0.31 |
TMEM163 |
transmembrane protein 163 |
43182 |
0.2 |
| chr6_34250869_34251020 | 0.31 |
C6orf1 |
chromosome 6 open reading frame 1 |
33697 |
0.16 |
| chr1_154464906_154465057 | 0.31 |
SHE |
Src homology 2 domain containing E |
3339 |
0.16 |
| chr5_39155242_39155486 | 0.31 |
FYB |
FYN binding protein |
47765 |
0.15 |
| chr3_50636542_50636693 | 0.31 |
CISH |
cytokine inducible SH2-containing protein |
12586 |
0.14 |
| chr16_67586263_67586449 | 0.31 |
FAM65A |
family with sequence similarity 65, member A |
8059 |
0.09 |
| chr5_10505521_10505706 | 0.31 |
RP11-1C1.5 |
Uncharacterized protein |
3693 |
0.21 |
| chr10_104830143_104830341 | 0.31 |
NT5C2 |
5'-nucleotidase, cytosolic II |
23883 |
0.21 |
| chr6_159454655_159454806 | 0.31 |
RP1-111C20.4 |
|
7752 |
0.19 |
| chr15_29228523_29228674 | 0.31 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
14758 |
0.23 |
| chr2_27109662_27109955 | 0.31 |
DPYSL5 |
dihydropyrimidinase-like 5 |
38432 |
0.13 |
| chr16_57175250_57175426 | 0.31 |
CPNE2 |
copine II |
22227 |
0.12 |
| chr4_124538385_124538719 | 0.31 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
217429 |
0.02 |
| chr9_117144402_117144633 | 0.31 |
AKNA |
AT-hook transcription factor |
4602 |
0.23 |
| chr7_50357344_50357742 | 0.31 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
9225 |
0.29 |
| chr18_60198457_60198694 | 0.31 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
7723 |
0.28 |
| chr1_47654400_47654551 | 0.31 |
PDZK1IP1 |
PDZK1 interacting protein 1 |
1252 |
0.4 |
| chr2_8519563_8519714 | 0.31 |
AC011747.7 |
|
296258 |
0.01 |
| chr22_39296706_39296951 | 0.31 |
CBX6 |
chromobox homolog 6 |
28509 |
0.12 |
| chr3_46416206_46416357 | 0.30 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
2120 |
0.26 |
| chr22_40798855_40799170 | 0.30 |
SGSM3 |
small G protein signaling modulator 3 |
5305 |
0.17 |
| chr1_167570221_167570504 | 0.30 |
RCSD1 |
RCSD domain containing 1 |
28968 |
0.15 |
| chr19_49838163_49838543 | 0.30 |
CD37 |
CD37 molecule |
300 |
0.78 |
| chr19_7402225_7402456 | 0.30 |
CTB-133G6.1 |
|
11508 |
0.16 |
| chr11_44936863_44937276 | 0.30 |
TSPAN18 |
tetraspanin 18 |
9111 |
0.25 |
| chr6_37485958_37486109 | 0.30 |
CCDC167 |
coiled-coil domain containing 167 |
18335 |
0.19 |
| chr2_8716699_8716875 | 0.30 |
AC011747.7 |
|
99109 |
0.08 |
| chr2_158278417_158278728 | 0.30 |
CYTIP |
cytohesin 1 interacting protein |
17354 |
0.21 |
| chr13_110222272_110222771 | 0.29 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
158108 |
0.04 |
| chr1_12233372_12233638 | 0.29 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
6445 |
0.17 |
| chr14_93533102_93533332 | 0.29 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
514 |
0.5 |
| chr8_24134022_24134173 | 0.29 |
ADAM28 |
ADAM metallopeptidase domain 28 |
17456 |
0.26 |
| chr8_6584199_6584352 | 0.29 |
AGPAT5 |
1-acylglycerol-3-phosphate O-acyltransferase 5 |
18061 |
0.17 |
| chr5_53912958_53913109 | 0.29 |
SNX18 |
sorting nexin 18 |
99440 |
0.08 |
| chr1_198529109_198529554 | 0.29 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
19256 |
0.26 |
| chr4_2837237_2837388 | 0.29 |
ADD1 |
adducin 1 (alpha) |
8272 |
0.19 |
| chr2_16105641_16106067 | 0.29 |
ENSG00000243541 |
. |
14751 |
0.18 |
| chr22_45028998_45029149 | 0.29 |
PRR5 |
proline rich 5 (renal) |
35520 |
0.18 |
| chr3_178793021_178793370 | 0.29 |
ZMAT3 |
zinc finger, matrin-type 3 |
3128 |
0.27 |
| chr14_61911054_61911390 | 0.29 |
PRKCH |
protein kinase C, eta |
1946 |
0.42 |
| chr17_76779813_76780089 | 0.29 |
CYTH1 |
cytohesin 1 |
1572 |
0.38 |
| chr7_73630981_73631230 | 0.29 |
LAT2 |
linker for activation of T cells family, member 2 |
6752 |
0.16 |
| chr1_247336048_247336203 | 0.29 |
ZNF124 |
zinc finger protein 124 |
807 |
0.56 |
| chr10_73494821_73495244 | 0.29 |
C10orf105 |
chromosome 10 open reading frame 105 |
2549 |
0.3 |
| chr22_24058094_24058402 | 0.29 |
AP000347.2 |
|
1234 |
0.32 |
| chr2_43610834_43610985 | 0.29 |
ENSG00000252804 |
. |
24537 |
0.25 |
| chr22_23134104_23134282 | 0.28 |
ENSG00000207833 |
. |
784 |
0.23 |
| chr9_132658123_132658578 | 0.28 |
FNBP1 |
formin binding protein 1 |
23239 |
0.15 |
| chr20_39780023_39780174 | 0.28 |
RP1-1J6.2 |
|
13455 |
0.19 |
| chr11_3969138_3969471 | 0.28 |
STIM1 |
stromal interaction molecule 1 |
731 |
0.66 |
| chr5_75720501_75720652 | 0.28 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
20327 |
0.26 |
| chr11_48039113_48039300 | 0.28 |
AC103828.1 |
|
1799 |
0.39 |
| chr5_171333279_171333614 | 0.28 |
FBXW11 |
F-box and WD repeat domain containing 11 |
71314 |
0.11 |
| chr1_227936240_227936391 | 0.28 |
SNAP47 |
synaptosomal-associated protein, 47kDa |
551 |
0.67 |
| chr2_198167309_198167571 | 0.28 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
197 |
0.93 |
| chr14_93134593_93134845 | 0.28 |
RIN3 |
Ras and Rab interactor 3 |
15873 |
0.24 |
| chr14_28295050_28295241 | 0.28 |
ENSG00000223164 |
. |
164066 |
0.04 |
| chr7_132118427_132118619 | 0.28 |
AC011625.1 |
|
81430 |
0.1 |
| chr17_74269143_74269364 | 0.28 |
QRICH2 |
glutamine rich 2 |
7185 |
0.13 |
| chr5_141282643_141282794 | 0.28 |
KIAA0141 |
KIAA0141 |
20655 |
0.14 |
| chr8_6287473_6287678 | 0.28 |
RP11-115C21.2 |
|
22912 |
0.21 |
| chr20_31111792_31112136 | 0.28 |
C20orf112 |
chromosome 20 open reading frame 112 |
3307 |
0.23 |
| chr14_21269173_21269324 | 0.28 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
1317 |
0.31 |
| chr10_7525377_7526075 | 0.28 |
ENSG00000207453 |
. |
3045 |
0.32 |
| chr4_185683459_185683704 | 0.28 |
ACSL1 |
acyl-CoA synthetase long-chain family member 1 |
4275 |
0.22 |
| chr1_100877624_100878071 | 0.28 |
ENSG00000216067 |
. |
33516 |
0.16 |
| chr8_134086193_134086434 | 0.28 |
SLA |
Src-like-adaptor |
13710 |
0.23 |
| chr6_53209989_53210148 | 0.27 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
3519 |
0.27 |
| chr17_7454664_7454994 | 0.27 |
TNFSF12 |
tumor necrosis factor (ligand) superfamily, member 12 |
2358 |
0.08 |
| chr17_63022313_63022486 | 0.27 |
RP11-583F2.5 |
|
25628 |
0.14 |
| chr5_118619331_118619759 | 0.27 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
15096 |
0.18 |
| chr3_43298637_43298793 | 0.27 |
SNRK |
SNF related kinase |
29289 |
0.16 |
| chr15_91228796_91228947 | 0.27 |
RP11-387D10.3 |
|
24901 |
0.13 |
| chr14_61827530_61827767 | 0.27 |
PRKCH |
protein kinase C, eta |
144 |
0.97 |
| chr1_9486742_9486893 | 0.27 |
ENSG00000252956 |
. |
11020 |
0.24 |
| chr20_48969794_48969957 | 0.27 |
ENSG00000244376 |
. |
76145 |
0.1 |
| chr14_91544790_91544971 | 0.27 |
ENSG00000252644 |
. |
12541 |
0.16 |
| chr3_141989336_141989501 | 0.27 |
GK5 |
glycerol kinase 5 (putative) |
44969 |
0.16 |
| chr13_114817282_114817433 | 0.27 |
RASA3 |
RAS p21 protein activator 3 |
26081 |
0.22 |
| chr16_3627581_3627750 | 0.27 |
NLRC3 |
NLR family, CARD domain containing 3 |
264 |
0.89 |
| chr16_30392632_30392982 | 0.27 |
SEPT1 |
septin 1 |
945 |
0.31 |
| chr12_132663008_132663169 | 0.27 |
GALNT9 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9) |
27249 |
0.14 |
| chr16_50196893_50197200 | 0.27 |
PAPD5 |
PAP associated domain containing 5 |
9267 |
0.16 |
| chr9_101900893_101901046 | 0.27 |
ENSG00000252942 |
. |
7279 |
0.19 |
| chr9_2023792_2024048 | 0.27 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
1975 |
0.41 |
| chr19_8632582_8633028 | 0.27 |
MYO1F |
myosin IF |
9517 |
0.11 |
| chr7_37461432_37461682 | 0.27 |
ENSG00000200113 |
. |
2326 |
0.3 |
| chr14_105750101_105750417 | 0.27 |
BRF1 |
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
16546 |
0.15 |
| chr19_17514773_17515020 | 0.27 |
BST2 |
bone marrow stromal cell antigen 2 |
1561 |
0.17 |
| chr22_23164320_23164502 | 0.26 |
IGLV2-8 |
immunoglobulin lambda variable 2-8 |
742 |
0.25 |
| chr6_158306516_158306953 | 0.26 |
RP3-403L10.3 |
|
7084 |
0.18 |
| chr7_7963215_7963560 | 0.26 |
ENSG00000201747 |
. |
17237 |
0.17 |
| chr14_22964920_22965173 | 0.26 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
8875 |
0.1 |
| chr1_206733046_206733294 | 0.26 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
2677 |
0.24 |
| chr22_39920591_39920772 | 0.26 |
ATF4 |
activating transcription factor 4 |
4112 |
0.17 |
| chr1_55668824_55669013 | 0.26 |
USP24 |
ubiquitin specific peptidase 24 |
11844 |
0.22 |
| chr6_109770732_109770883 | 0.26 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
179 |
0.91 |
| chr1_167452877_167453134 | 0.26 |
RP11-104L21.2 |
|
25107 |
0.17 |
| chr7_139468099_139468250 | 0.26 |
HIPK2 |
homeodomain interacting protein kinase 2 |
9343 |
0.23 |
| chr11_48065545_48066080 | 0.26 |
AC103828.1 |
|
28405 |
0.17 |
| chr17_72636963_72637310 | 0.26 |
CD300E |
CD300e molecule |
17239 |
0.12 |
| chr5_142178337_142178630 | 0.26 |
ARHGAP26 |
Rho GTPase activating protein 26 |
26201 |
0.22 |
| chr6_106324555_106324706 | 0.26 |
ENSG00000200198 |
. |
27618 |
0.26 |
| chr3_34060384_34060651 | 0.26 |
PDCD6IP |
programmed cell death 6 interacting protein |
220402 |
0.02 |
| chr1_247367941_247368092 | 0.26 |
ENSG00000252516 |
. |
53 |
0.96 |
| chr8_71053263_71053636 | 0.26 |
NCOA2 |
nuclear receptor coactivator 2 |
7172 |
0.25 |
| chr5_67527341_67527547 | 0.26 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
4982 |
0.28 |
| chr12_108714499_108714650 | 0.26 |
CMKLR1 |
chemokine-like receptor 1 |
135 |
0.98 |
| chr6_143865508_143865659 | 0.26 |
PHACTR2 |
phosphatase and actin regulator 2 |
7601 |
0.17 |
| chr18_3723612_3723763 | 0.25 |
RP11-874J12.3 |
|
47743 |
0.13 |
| chr18_43589850_43590001 | 0.25 |
ENSG00000222179 |
. |
20014 |
0.17 |
| chr1_154987014_154987262 | 0.25 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
214 |
0.83 |
| chr18_21549060_21549211 | 0.25 |
LAMA3 |
laminin, alpha 3 |
19278 |
0.17 |
| chr8_37553639_37554052 | 0.25 |
ZNF703 |
zinc finger protein 703 |
576 |
0.66 |
| chr6_157140981_157141263 | 0.25 |
RP11-230C9.3 |
|
39533 |
0.16 |
| chr16_31497874_31498025 | 0.25 |
AC026471.6 |
|
1864 |
0.17 |
| chr1_172694568_172694750 | 0.25 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
66501 |
0.13 |
| chr19_12442067_12442220 | 0.25 |
ZNF563 |
zinc finger protein 563 |
2163 |
0.24 |
| chr22_43659141_43659490 | 0.25 |
SCUBE1 |
signal peptide, CUB domain, EGF-like 1 |
510 |
0.76 |
| chr1_86907105_86907371 | 0.25 |
CLCA2 |
chloride channel accessory 2 |
17469 |
0.19 |
| chr1_66773813_66773964 | 0.25 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
22523 |
0.27 |
| chr1_100830416_100830661 | 0.25 |
CDC14A |
cell division cycle 14A |
12033 |
0.18 |
| chr13_84712610_84712776 | 0.25 |
SLITRK1 |
SLIT and NTRK-like family, member 1 |
256165 |
0.02 |
| chr5_176859524_176860060 | 0.25 |
GRK6 |
G protein-coupled receptor kinase 6 |
5937 |
0.1 |
| chr15_31728916_31729103 | 0.25 |
KLF13 |
Kruppel-like factor 13 |
70652 |
0.13 |
| chr2_218884404_218884588 | 0.25 |
TNS1 |
tensin 1 |
16778 |
0.16 |
| chr20_55097345_55097496 | 0.25 |
FAM209A |
family with sequence similarity 209, member A |
2122 |
0.22 |
| chr17_26070911_26071062 | 0.25 |
ENSG00000263472 |
. |
41048 |
0.13 |
| chr9_95822386_95822880 | 0.25 |
SUSD3 |
sushi domain containing 3 |
1572 |
0.37 |
| chr1_112887579_112887730 | 0.25 |
ENSG00000238761 |
. |
25972 |
0.2 |
| chr1_1074452_1074741 | 0.25 |
C1orf159 |
chromosome 1 open reading frame 159 |
22855 |
0.07 |
| chr4_2789296_2789952 | 0.25 |
SH3BP2 |
SH3-domain binding protein 2 |
5126 |
0.22 |
| chr2_103511305_103511456 | 0.25 |
TMEM182 |
transmembrane protein 182 |
132908 |
0.05 |
| chr10_91498741_91498892 | 0.25 |
KIF20B |
kinesin family member 20B |
37380 |
0.18 |
| chr2_99090097_99090281 | 0.25 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
28776 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.3 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 0.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.3 | GO:0045588 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.1 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 | 0.2 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.5 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.1 | 0.3 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.2 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.1 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0002664 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.4 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0002448 | mast cell mediated immunity(GO:0002448) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.4 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.6 | GO:0002639 | positive regulation of immunoglobulin production(GO:0002639) |
| 0.0 | 0.2 | GO:0001562 | response to protozoan(GO:0001562) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.4 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.3 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0071635 | negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.2 | GO:0032367 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) mucosal immune response(GO:0002385) |
| 0.0 | 0.4 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0097300 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.2 | GO:0051647 | nucleus localization(GO:0051647) |
| 0.0 | 0.1 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.2 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.2 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0010523 | negative regulation of calcium ion transport into cytosol(GO:0010523) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.2 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.9 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0070723 | response to sterol(GO:0036314) response to cholesterol(GO:0070723) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.0 | GO:0090286 | nuclear matrix organization(GO:0043578) cytoskeletal anchoring at nuclear membrane(GO:0090286) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.0 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.1 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0002251 | organ or tissue specific immune response(GO:0002251) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.1 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.0 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0031063 | regulation of histone deacetylation(GO:0031063) positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.0 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.0 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.2 | GO:0070192 | synapsis(GO:0007129) chromosome organization involved in meiotic cell cycle(GO:0070192) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0060557 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.0 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.0 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.0 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) |
| 0.0 | 0.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0046015 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:1903224 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.0 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.0 | GO:0097384 | cellular lipid biosynthetic process(GO:0097384) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.0 | GO:0031058 | positive regulation of histone modification(GO:0031058) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.0 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.0 | GO:0051818 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.1 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.0 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0042597 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.4 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.0 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.1 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.8 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.3 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.0 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.0 | 0.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |