Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
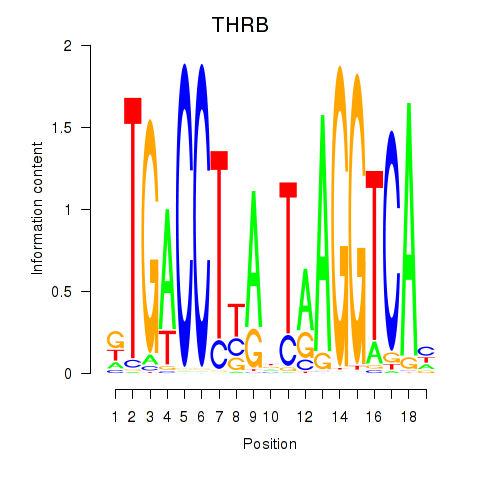
Results for THRB
Z-value: 0.95
Transcription factors associated with THRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRB
|
ENSG00000151090.13 | thyroid hormone receptor beta |
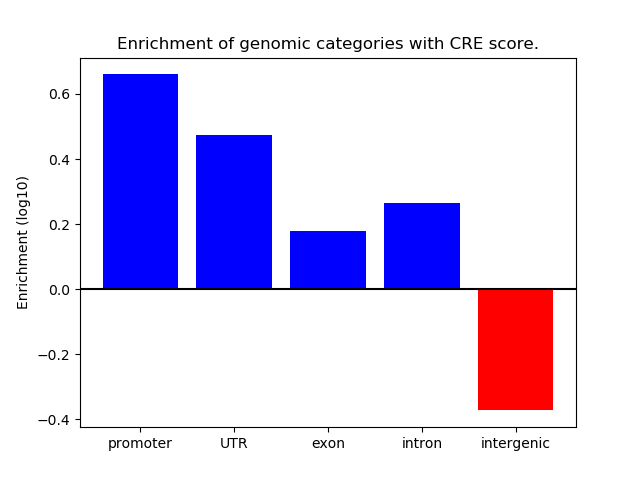
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_24193778_24193929 | THRB | 13233 | 0.292606 | -0.80 | 9.9e-03 | Click! |
| chr3_24283925_24284076 | THRB | 76914 | 0.115963 | 0.77 | 1.4e-02 | Click! |
| chr3_24536950_24537123 | THRB | 263 | 0.882887 | -0.72 | 2.8e-02 | Click! |
| chr3_24271931_24272082 | THRB | 64920 | 0.141320 | -0.65 | 5.6e-02 | Click! |
| chr3_24187341_24187492 | THRB | 19670 | 0.273525 | 0.63 | 7.0e-02 | Click! |
Activity of the THRB motif across conditions
Conditions sorted by the z-value of the THRB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
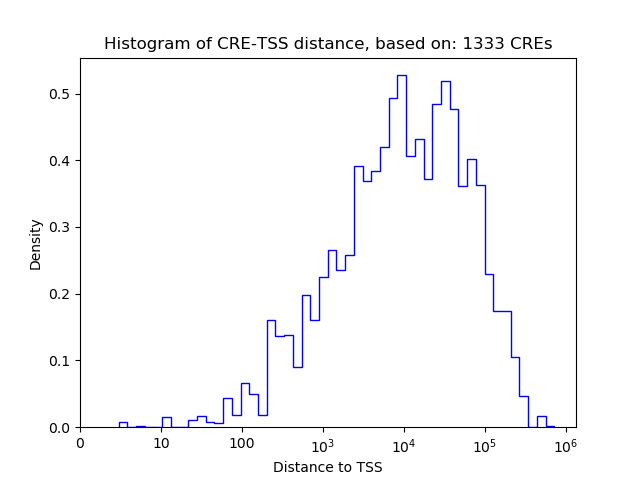
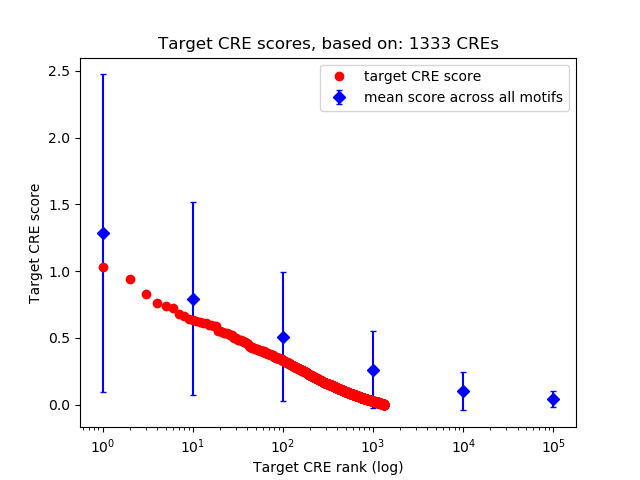
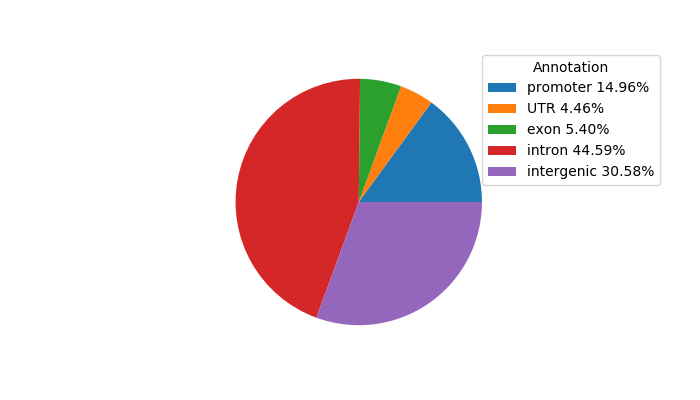
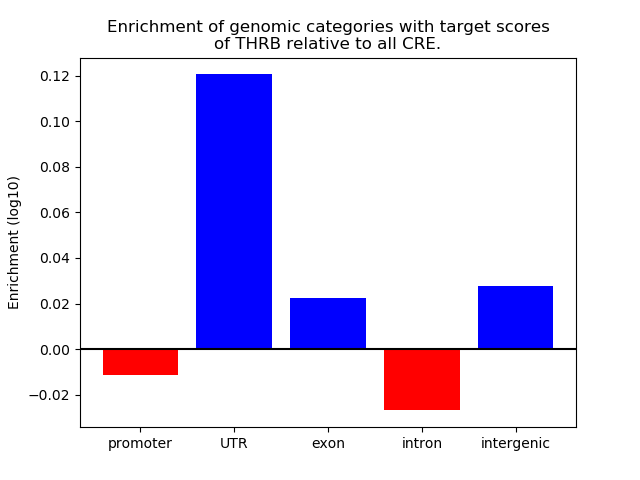
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_123429411_123429620 | 1.03 |
ABCB9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
293 |
0.88 |
| chr17_2869363_2869514 | 0.94 |
CTD-3060P21.1 |
|
249 |
0.94 |
| chr20_49983594_49983745 | 0.83 |
ENSG00000263645 |
. |
10189 |
0.28 |
| chr17_70404089_70404240 | 0.76 |
ENSG00000200783 |
. |
256127 |
0.02 |
| chr2_175307874_175308025 | 0.74 |
GPR155 |
G protein-coupled receptor 155 |
43816 |
0.12 |
| chr8_18991967_18992118 | 0.73 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
49802 |
0.18 |
| chr2_228735932_228736236 | 0.68 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
239 |
0.94 |
| chr17_77023771_77023922 | 0.67 |
C1QTNF1-AS1 |
C1QTNF1 antisense RNA 1 |
109 |
0.95 |
| chr2_36635278_36635429 | 0.64 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
51739 |
0.16 |
| chr1_27864005_27864199 | 0.64 |
RP1-159A19.4 |
|
11786 |
0.17 |
| chr8_30656188_30656339 | 0.63 |
PPP2CB |
protein phosphatase 2, catalytic subunit, beta isozyme |
4466 |
0.26 |
| chr1_86021034_86021185 | 0.62 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
22824 |
0.17 |
| chr17_58966579_58966730 | 0.61 |
BCAS3 |
breast carcinoma amplified sequence 3 |
2014 |
0.33 |
| chr5_52655964_52656115 | 0.61 |
FST |
follistatin |
120200 |
0.06 |
| chr1_7531258_7531628 | 0.60 |
RP4-549F15.1 |
|
29829 |
0.21 |
| chr5_111017081_111017232 | 0.60 |
ENSG00000253057 |
. |
38602 |
0.16 |
| chr12_76256655_76256806 | 0.59 |
ENSG00000243420 |
. |
95180 |
0.08 |
| chr1_204141000_204141151 | 0.59 |
REN |
renin |
5610 |
0.15 |
| chr10_81915037_81915188 | 0.55 |
ANXA11 |
annexin A11 |
6656 |
0.2 |
| chr5_159339303_159339454 | 0.55 |
ADRA1B |
adrenoceptor alpha 1B |
4412 |
0.31 |
| chr15_68911339_68911490 | 0.55 |
CORO2B |
coronin, actin binding protein, 2B |
2535 |
0.41 |
| chr9_110816490_110816641 | 0.54 |
ENSG00000222459 |
. |
135306 |
0.05 |
| chr16_81601760_81601924 | 0.54 |
CMIP |
c-Maf inducing protein |
72888 |
0.12 |
| chr8_22454417_22454792 | 0.54 |
C8orf58 |
chromosome 8 open reading frame 58 |
2510 |
0.15 |
| chr15_83223362_83223513 | 0.53 |
RP11-152F13.10 |
|
1245 |
0.33 |
| chr17_80846919_80847070 | 0.52 |
TBCD |
tubulin folding cofactor D |
3034 |
0.26 |
| chr21_30047800_30047951 | 0.52 |
ENSG00000251894 |
. |
67655 |
0.14 |
| chr16_27419960_27420215 | 0.51 |
IL21R |
interleukin 21 receptor |
5664 |
0.22 |
| chr9_132399053_132399204 | 0.50 |
RP11-483H20.4 |
|
3627 |
0.14 |
| chr2_10592171_10592322 | 0.50 |
ODC1 |
ornithine decarboxylase 1 |
3616 |
0.22 |
| chr15_31653474_31653647 | 0.49 |
KLF13 |
Kruppel-like factor 13 |
4797 |
0.34 |
| chr16_12214413_12214696 | 0.49 |
SNX29 |
sorting nexin 29 |
9000 |
0.18 |
| chr9_110947632_110947783 | 0.48 |
ENSG00000222512 |
. |
173502 |
0.03 |
| chr14_24111076_24111295 | 0.48 |
DHRS2 |
dehydrogenase/reductase (SDR family) member 2 |
2637 |
0.24 |
| chr1_218833924_218834075 | 0.48 |
ENSG00000212610 |
. |
118758 |
0.06 |
| chr7_55123506_55123657 | 0.48 |
EGFR |
epidermal growth factor receptor |
36770 |
0.22 |
| chr7_157064029_157064180 | 0.47 |
ENSG00000266453 |
. |
34383 |
0.19 |
| chr17_1639800_1639951 | 0.47 |
RP11-961A15.1 |
|
2004 |
0.18 |
| chr21_46432509_46432660 | 0.46 |
AP001579.1 |
Uncharacterized protein |
60343 |
0.07 |
| chr1_236107074_236107261 | 0.46 |
ENSG00000206803 |
. |
28452 |
0.17 |
| chr7_2548232_2548731 | 0.46 |
LFNG |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
3682 |
0.2 |
| chr5_72672425_72672576 | 0.44 |
FOXD1 |
forkhead box D1 |
71852 |
0.1 |
| chr4_169408008_169408159 | 0.44 |
DDX60L |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
6418 |
0.2 |
| chr11_10314214_10314808 | 0.43 |
SBF2 |
SET binding factor 2 |
1243 |
0.42 |
| chr10_92066348_92066499 | 0.43 |
ENSG00000222451 |
. |
142706 |
0.05 |
| chr3_100058113_100058264 | 0.43 |
NIT2 |
nitrilase family, member 2 |
4533 |
0.24 |
| chr1_61914926_61915077 | 0.43 |
NFIA |
nuclear factor I/A |
42657 |
0.21 |
| chr17_48141828_48141980 | 0.42 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
6333 |
0.14 |
| chr15_67316146_67316685 | 0.42 |
SMAD3 |
SMAD family member 3 |
39686 |
0.2 |
| chr6_7671378_7671529 | 0.42 |
BMP6 |
bone morphogenetic protein 6 |
55577 |
0.14 |
| chr8_32775609_32775811 | 0.42 |
ENSG00000212407 |
. |
6594 |
0.33 |
| chr5_149844207_149844358 | 0.41 |
RPS14 |
ribosomal protein S14 |
14963 |
0.16 |
| chr14_62030492_62030755 | 0.41 |
RP11-47I22.3 |
Uncharacterized protein |
6691 |
0.21 |
| chr2_75292380_75292531 | 0.41 |
ENSG00000263909 |
. |
25484 |
0.21 |
| chr3_20098327_20098803 | 0.41 |
KAT2B |
K(lysine) acetyltransferase 2B |
17050 |
0.18 |
| chr2_122015503_122015654 | 0.40 |
TFCP2L1 |
transcription factor CP2-like 1 |
27205 |
0.25 |
| chr12_52674373_52674524 | 0.40 |
RP11-845M18.7 |
|
1508 |
0.22 |
| chr7_214681_214832 | 0.40 |
AC145676.2 |
Uncharacterized protein |
14801 |
0.19 |
| chr10_49904377_49904528 | 0.40 |
WDFY4 |
WDFY family member 4 |
11531 |
0.22 |
| chr22_20887761_20888231 | 0.40 |
MED15 |
mediator complex subunit 15 |
9434 |
0.12 |
| chr6_36644830_36645109 | 0.40 |
CDKN1A |
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
619 |
0.65 |
| chr22_36445355_36445635 | 0.40 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
21022 |
0.24 |
| chr1_226849410_226849624 | 0.39 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
2146 |
0.33 |
| chr11_64427589_64427881 | 0.39 |
AP001092.4 |
|
13864 |
0.15 |
| chr6_169572510_169572661 | 0.39 |
XXyac-YX65C7_A.2 |
|
40764 |
0.2 |
| chr3_122486711_122486862 | 0.39 |
ENSG00000238480 |
. |
17567 |
0.17 |
| chr17_45343433_45343839 | 0.39 |
ENSG00000238419 |
. |
6850 |
0.14 |
| chr11_122052864_122053015 | 0.38 |
ENSG00000207994 |
. |
29923 |
0.15 |
| chr1_169083123_169083274 | 0.38 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
3362 |
0.23 |
| chr3_189433263_189433414 | 0.38 |
TP63 |
tumor protein p63 |
74111 |
0.12 |
| chr11_2224293_2224574 | 0.38 |
ENSG00000265258 |
. |
30140 |
0.1 |
| chr9_5839505_5839749 | 0.38 |
ERMP1 |
endoplasmic reticulum metallopeptidase 1 |
6510 |
0.22 |
| chr22_38749450_38749601 | 0.38 |
RP3-449O17.1 |
|
30576 |
0.11 |
| chr19_55899777_55900128 | 0.37 |
RPL28 |
ribosomal protein L28 |
2226 |
0.12 |
| chr12_26351604_26351755 | 0.37 |
SSPN |
sarcospan |
3073 |
0.29 |
| chr9_107816671_107816822 | 0.37 |
ENSG00000201583 |
. |
42291 |
0.19 |
| chr4_148736144_148736295 | 0.37 |
ENSG00000240014 |
. |
6142 |
0.23 |
| chr12_95189281_95189432 | 0.36 |
ENSG00000208038 |
. |
38818 |
0.2 |
| chr12_112550183_112550334 | 0.36 |
NAA25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
3432 |
0.18 |
| chr1_114000056_114000207 | 0.36 |
MAGI3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
66520 |
0.14 |
| chr16_20952479_20952845 | 0.36 |
LYRM1 |
LYR motif containing 1 |
40236 |
0.13 |
| chr2_62881644_62881884 | 0.35 |
AC092155.4 |
|
7999 |
0.23 |
| chr12_66350099_66350285 | 0.35 |
RP11-366L20.3 |
|
7933 |
0.21 |
| chr15_91318037_91318188 | 0.35 |
ENSG00000200677 |
. |
19570 |
0.14 |
| chr2_65543802_65543953 | 0.35 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
18030 |
0.23 |
| chr12_6338368_6338538 | 0.35 |
ENSG00000202318 |
. |
5168 |
0.2 |
| chr17_62317609_62317841 | 0.35 |
TEX2 |
testis expressed 2 |
9609 |
0.22 |
| chr4_170053647_170053798 | 0.35 |
RP11-327O17.2 |
|
69223 |
0.11 |
| chr11_12753644_12753795 | 0.35 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
12869 |
0.26 |
| chr14_103868479_103868639 | 0.35 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
15983 |
0.16 |
| chr7_121184730_121184881 | 0.35 |
ENSG00000221690 |
. |
30003 |
0.24 |
| chr9_132249416_132249920 | 0.35 |
ENSG00000264298 |
. |
8833 |
0.22 |
| chr14_92864210_92864675 | 0.34 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
10281 |
0.28 |
| chr2_14773054_14773337 | 0.34 |
FAM84A |
family with sequence similarity 84, member A |
371 |
0.88 |
| chr5_112483190_112483341 | 0.34 |
MCC |
mutated in colorectal cancers |
87109 |
0.09 |
| chr18_3653050_3653341 | 0.34 |
RP11-874J12.4 |
|
215 |
0.94 |
| chr14_23308927_23309078 | 0.34 |
MMP14 |
matrix metallopeptidase 14 (membrane-inserted) |
2481 |
0.13 |
| chr2_169378165_169378347 | 0.34 |
ENSG00000265694 |
. |
61197 |
0.1 |
| chr1_234722064_234722215 | 0.34 |
ENSG00000212144 |
. |
6882 |
0.2 |
| chr3_50606139_50606449 | 0.33 |
HEMK1 |
HemK methyltransferase family member 1 |
289 |
0.82 |
| chr3_185482052_185482385 | 0.33 |
ENSG00000265470 |
. |
3474 |
0.28 |
| chr10_105610481_105610681 | 0.33 |
SH3PXD2A |
SH3 and PX domains 2A |
4583 |
0.23 |
| chr4_71875471_71875927 | 0.33 |
DCK |
deoxycytidine kinase |
16344 |
0.25 |
| chr9_35258666_35258853 | 0.33 |
UNC13B |
unc-13 homolog B (C. elegans) |
96658 |
0.06 |
| chr1_92202773_92202924 | 0.32 |
ENSG00000239794 |
. |
92783 |
0.08 |
| chr8_32407810_32407961 | 0.32 |
NRG1 |
neuregulin 1 |
1640 |
0.54 |
| chr14_22693388_22693539 | 0.32 |
ENSG00000238634 |
. |
82576 |
0.09 |
| chr9_132949422_132949573 | 0.32 |
NCS1 |
neuronal calcium sensor 1 |
13375 |
0.21 |
| chr9_22237739_22238051 | 0.32 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
124218 |
0.06 |
| chr11_61748663_61748814 | 0.32 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
13285 |
0.13 |
| chr17_73872544_73873300 | 0.32 |
TRIM47 |
tripartite motif containing 47 |
423 |
0.43 |
| chr14_105145297_105145448 | 0.31 |
ENSG00000265291 |
. |
1286 |
0.36 |
| chr1_218649534_218649685 | 0.31 |
C1orf143 |
chromosome 1 open reading frame 143 |
33829 |
0.2 |
| chr18_24127643_24127794 | 0.31 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
636 |
0.79 |
| chr3_124879753_124879904 | 0.31 |
ENSG00000264986 |
. |
9432 |
0.2 |
| chr2_173116908_173117059 | 0.31 |
ENSG00000238572 |
. |
96135 |
0.07 |
| chr17_29014178_29014503 | 0.31 |
ENSG00000241631 |
. |
15453 |
0.16 |
| chr22_30194899_30195349 | 0.31 |
ASCC2 |
activating signal cointegrator 1 complex subunit 2 |
2967 |
0.2 |
| chr13_40738441_40738592 | 0.30 |
ENSG00000207458 |
. |
62448 |
0.15 |
| chr2_216769053_216769204 | 0.30 |
ENSG00000212055 |
. |
25486 |
0.24 |
| chr4_174293358_174293974 | 0.30 |
SAP30 |
Sin3A-associated protein, 30kDa |
1572 |
0.28 |
| chr13_49081098_49081249 | 0.30 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
26040 |
0.23 |
| chr1_27667588_27668215 | 0.30 |
SYTL1 |
synaptotagmin-like 1 |
612 |
0.61 |
| chrX_31217215_31217366 | 0.30 |
DMD |
dystrophin |
7427 |
0.31 |
| chr17_77022601_77022884 | 0.30 |
C1QTNF1-AS1 |
C1QTNF1 antisense RNA 1 |
941 |
0.36 |
| chr22_30601714_30601935 | 0.30 |
RP3-438O4.4 |
|
1274 |
0.36 |
| chr8_97349873_97350371 | 0.30 |
ENSG00000199732 |
. |
28572 |
0.17 |
| chr17_21197723_21197874 | 0.29 |
MAP2K3 |
mitogen-activated protein kinase kinase 3 |
6440 |
0.2 |
| chr9_130906009_130906346 | 0.29 |
LCN2 |
lipocalin 2 |
5173 |
0.09 |
| chr10_72016101_72016252 | 0.29 |
PPA1 |
pyrophosphatase (inorganic) 1 |
22993 |
0.17 |
| chr3_99255664_99255815 | 0.29 |
COL8A1 |
collagen, type VIII, alpha 1 |
101580 |
0.08 |
| chr5_151058528_151058679 | 0.29 |
CTB-113P19.1 |
|
2069 |
0.25 |
| chr5_64689733_64689884 | 0.29 |
ADAMTS6 |
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
79710 |
0.1 |
| chr2_237677199_237677457 | 0.28 |
ACKR3 |
atypical chemokine receptor 3 |
199044 |
0.03 |
| chr16_89666849_89667000 | 0.28 |
CPNE7 |
copine VII |
4940 |
0.12 |
| chr3_172033208_172033359 | 0.28 |
AC092964.1 |
Uncharacterized protein |
935 |
0.62 |
| chr11_106328094_106328245 | 0.28 |
RP11-680E19.1 |
|
193127 |
0.03 |
| chr1_234752857_234753272 | 0.28 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
7793 |
0.19 |
| chr1_41162222_41162675 | 0.28 |
NFYC |
nuclear transcription factor Y, gamma |
4486 |
0.17 |
| chr22_23035689_23035840 | 0.28 |
IGLV3-24 |
immunoglobulin lambda variable 3-24 (pseudogene) |
1160 |
0.15 |
| chr2_73248670_73248821 | 0.28 |
SFXN5 |
sideroflexin 5 |
50057 |
0.13 |
| chr11_131920807_131920958 | 0.28 |
RP11-697E14.2 |
|
66327 |
0.14 |
| chr11_72500407_72500904 | 0.28 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
3643 |
0.14 |
| chr19_58893310_58893806 | 0.28 |
ZNF837 |
zinc finger protein 837 |
1131 |
0.22 |
| chr21_29625341_29625492 | 0.27 |
ENSG00000251894 |
. |
490114 |
0.01 |
| chr2_64979976_64980127 | 0.27 |
ENSG00000253082 |
. |
23187 |
0.19 |
| chr17_1520967_1521246 | 0.27 |
ENSG00000238946 |
. |
1517 |
0.25 |
| chr1_208358461_208358612 | 0.27 |
PLXNA2 |
plexin A2 |
59129 |
0.17 |
| chr2_29337664_29337897 | 0.27 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
512 |
0.76 |
| chrX_34919614_34919765 | 0.27 |
FAM47B |
family with sequence similarity 47, member B |
41224 |
0.22 |
| chr4_2802460_2802681 | 0.27 |
SH3BP2 |
SH3-domain binding protein 2 |
1845 |
0.36 |
| chr5_106695662_106695882 | 0.27 |
EFNA5 |
ephrin-A5 |
310556 |
0.01 |
| chr21_39648260_39648411 | 0.27 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
3906 |
0.33 |
| chr5_142885689_142885840 | 0.27 |
ENSG00000253023 |
. |
36388 |
0.19 |
| chr9_119713049_119713301 | 0.27 |
ENSG00000265662 |
. |
148111 |
0.04 |
| chr17_78644745_78644896 | 0.26 |
RPTOR |
regulatory associated protein of MTOR, complex 1 |
125552 |
0.05 |
| chr14_73268075_73268226 | 0.26 |
ENSG00000206751 |
. |
51889 |
0.14 |
| chr13_40810218_40810369 | 0.26 |
ENSG00000207458 |
. |
9329 |
0.31 |
| chr3_72536153_72536343 | 0.26 |
RYBP |
RING1 and YY1 binding protein |
40179 |
0.18 |
| chr2_74810778_74811272 | 0.26 |
LOXL3 |
lysyl oxidase-like 3 |
28208 |
0.08 |
| chr11_106269511_106269662 | 0.26 |
RP11-680E19.1 |
|
134544 |
0.05 |
| chr1_224920982_224921133 | 0.26 |
RP11-3L21.2 |
|
2693 |
0.27 |
| chr5_95630524_95630804 | 0.26 |
ENSG00000206997 |
. |
84733 |
0.09 |
| chr11_87064154_87064305 | 0.26 |
ENSG00000223015 |
. |
253446 |
0.02 |
| chr15_85457860_85458027 | 0.25 |
ENSG00000207037 |
. |
23810 |
0.13 |
| chrX_154297234_154297385 | 0.25 |
MTCP1 |
mature T-cell proliferation 1 |
2192 |
0.2 |
| chr4_38077270_38077549 | 0.25 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
29917 |
0.24 |
| chr11_12147970_12148167 | 0.25 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
5046 |
0.31 |
| chr1_12483776_12483927 | 0.25 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
13927 |
0.23 |
| chr15_41058349_41058500 | 0.25 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
606 |
0.58 |
| chr11_68351182_68351333 | 0.25 |
PPP6R3 |
protein phosphatase 6, regulatory subunit 3 |
25147 |
0.17 |
| chr20_48760575_48761068 | 0.25 |
TMEM189-UBE2V1 |
TMEM189-UBE2V1 readthrough |
9353 |
0.17 |
| chr7_105410894_105411045 | 0.25 |
ATXN7L1 |
ataxin 7-like 1 |
78885 |
0.11 |
| chr5_71762160_71762360 | 0.25 |
RP11-389C8.2 |
|
24046 |
0.21 |
| chr3_126340455_126340606 | 0.25 |
TXNRD3 |
thioredoxin reductase 3 |
12442 |
0.17 |
| chr14_100222322_100222621 | 0.25 |
EML1 |
echinoderm microtubule associated protein like 1 |
16815 |
0.21 |
| chr8_117049888_117050039 | 0.24 |
ENSG00000199450 |
. |
14959 |
0.29 |
| chr1_109704266_109704907 | 0.24 |
ENSG00000238310 |
. |
45884 |
0.08 |
| chr19_3434582_3434733 | 0.24 |
C19orf77 |
chromosome 19 open reading frame 77 |
43829 |
0.09 |
| chr3_184300499_184300650 | 0.24 |
EPHB3 |
EPH receptor B3 |
21002 |
0.18 |
| chr19_1365463_1365614 | 0.24 |
MUM1 |
melanoma associated antigen (mutated) 1 |
9175 |
0.08 |
| chr2_201391804_201392087 | 0.24 |
SGOL2 |
shugoshin-like 2 (S. pombe) |
1055 |
0.54 |
| chr11_5617899_5618367 | 0.24 |
TRIM6 |
tripartite motif containing 6 |
97 |
0.61 |
| chr12_27717712_27717863 | 0.24 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
26731 |
0.2 |
| chr4_69138479_69138630 | 0.24 |
TMPRSS11B |
transmembrane protease, serine 11B |
27116 |
0.16 |
| chr2_238223705_238223856 | 0.23 |
AC112715.2 |
Uncharacterized protein |
58046 |
0.13 |
| chr10_69848135_69848286 | 0.23 |
HERC4 |
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
13105 |
0.18 |
| chr9_21027016_21027377 | 0.23 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
4412 |
0.24 |
| chr20_25003716_25003867 | 0.23 |
ACSS1 |
acyl-CoA synthetase short-chain family member 1 |
9517 |
0.19 |
| chr22_36723096_36723607 | 0.23 |
ENSG00000266345 |
. |
11105 |
0.19 |
| chr4_129746477_129746767 | 0.23 |
JADE1 |
jade family PHD finger 1 |
5966 |
0.31 |
| chr9_131655343_131655643 | 0.23 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
10712 |
0.11 |
| chr4_88950180_88950331 | 0.23 |
PKD2 |
polycystic kidney disease 2 (autosomal dominant) |
14158 |
0.19 |
| chr17_18904649_18904946 | 0.23 |
FAM83G |
family with sequence similarity 83, member G |
2692 |
0.18 |
| chr17_46662373_46662833 | 0.23 |
HOXB3 |
homeobox B3 |
2619 |
0.1 |
| chr15_52371258_52371409 | 0.23 |
CTD-2184D3.5 |
|
21387 |
0.13 |
| chr11_1580523_1580674 | 0.22 |
DUSP8 |
dual specificity phosphatase 8 |
6568 |
0.13 |
| chr7_55141300_55141512 | 0.22 |
EGFR |
epidermal growth factor receptor |
36010 |
0.22 |
| chr4_187622907_187623058 | 0.22 |
FAT1 |
FAT atypical cadherin 1 |
22027 |
0.25 |
| chr17_7344055_7344232 | 0.22 |
FGF11 |
fibroblast growth factor 11 |
81 |
0.89 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.1 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.1 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.2 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.0 | GO:0051140 | regulation of NK T cell activation(GO:0051133) positive regulation of NK T cell activation(GO:0051135) regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.2 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0032048 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0090030 | regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:1903170 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0051571 | positive regulation of histone methylation(GO:0031062) positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0060177 | regulation of angiotensin levels in blood(GO:0002002) regulation of angiotensin metabolic process(GO:0060177) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.2 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.1 | GO:1901222 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.0 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |