Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
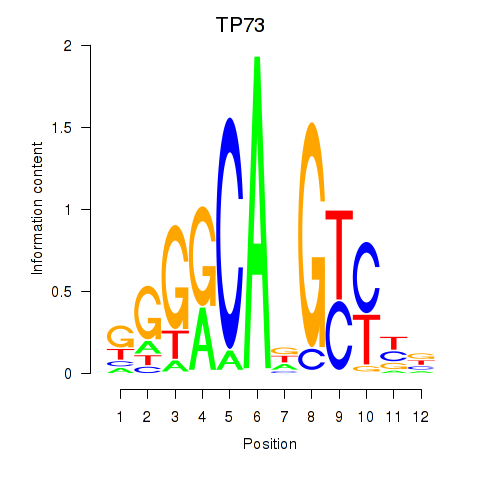
Results for TP73
Z-value: 0.93
Transcription factors associated with TP73
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TP73
|
ENSG00000078900.10 | tumor protein p73 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_3594845_3595177 | TP73 | 3919 | 0.141799 | -0.75 | 1.9e-02 | Click! |
| chr1_3593861_3594210 | TP73 | 4895 | 0.131181 | -0.65 | 5.7e-02 | Click! |
| chr1_3619704_3619855 | TP73 | 5157 | 0.128438 | 0.63 | 6.9e-02 | Click! |
| chr1_3593293_3593464 | TP73 | 5552 | 0.126652 | -0.63 | 6.9e-02 | Click! |
| chr1_3619930_3620081 | TP73 | 5383 | 0.126935 | 0.57 | 1.1e-01 | Click! |
Activity of the TP73 motif across conditions
Conditions sorted by the z-value of the TP73 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
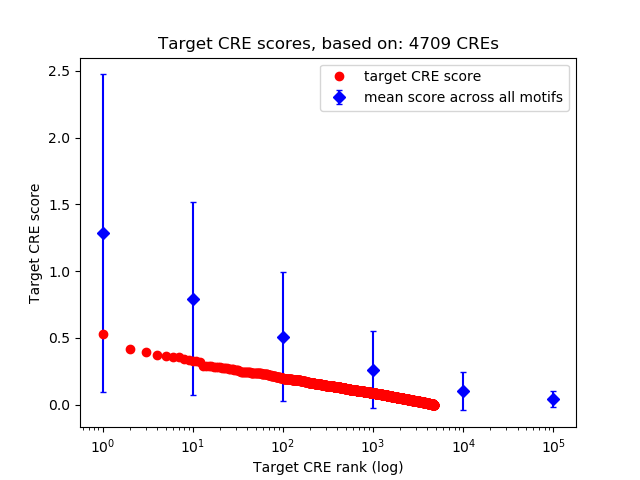
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_109833240_109833391 | 0.53 |
MYO16-AS1 |
MYO16 antisense RNA 1 |
20516 |
0.27 |
| chr3_133920629_133920780 | 0.42 |
RYK |
receptor-like tyrosine kinase |
18827 |
0.26 |
| chr15_74486059_74486210 | 0.40 |
STRA6 |
stimulated by retinoic acid 6 |
4560 |
0.14 |
| chr3_11254129_11254425 | 0.37 |
HRH1 |
histamine receptor H1 |
13440 |
0.26 |
| chr1_156211867_156212112 | 0.37 |
BGLAP |
bone gamma-carboxyglutamate (gla) protein |
236 |
0.85 |
| chr15_75995798_75996054 | 0.36 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
9263 |
0.1 |
| chr6_37045515_37045666 | 0.36 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
32983 |
0.16 |
| chr19_48991448_48991599 | 0.34 |
CYTH2 |
cytohesin 2 |
15914 |
0.09 |
| chr22_45949176_45949470 | 0.34 |
FBLN1 |
fibulin 1 |
34660 |
0.15 |
| chr15_78180472_78180634 | 0.33 |
CSPG4P13 |
chondroitin sulfate proteoglycan 4 pseudogene 13 |
6473 |
0.16 |
| chr22_43523288_43523514 | 0.33 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
15569 |
0.13 |
| chr16_27590487_27590638 | 0.32 |
KIAA0556 |
KIAA0556 |
5334 |
0.22 |
| chr9_137270522_137271255 | 0.29 |
ENSG00000263897 |
. |
369 |
0.9 |
| chr5_17187432_17187583 | 0.29 |
AC091878.1 |
|
29649 |
0.14 |
| chr1_19635741_19635892 | 0.29 |
AKR7A2 |
aldo-keto reductase family 7, member A2 (aflatoxin aldehyde reductase) |
2585 |
0.15 |
| chr12_52540643_52541115 | 0.29 |
ENSG00000265804 |
. |
33170 |
0.09 |
| chr16_8980256_8980407 | 0.29 |
RP11-77H9.6 |
|
17223 |
0.12 |
| chr5_73112011_73112162 | 0.28 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
2743 |
0.28 |
| chr9_125138658_125139005 | 0.28 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
1230 |
0.35 |
| chr15_90754517_90755171 | 0.28 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
10076 |
0.12 |
| chr22_18515057_18515208 | 0.28 |
MICAL3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
7807 |
0.15 |
| chr1_116325528_116325679 | 0.28 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
14201 |
0.24 |
| chr14_103577690_103577872 | 0.27 |
EXOC3L4 |
exocyst complex component 3-like 4 |
3928 |
0.19 |
| chr10_99306823_99306974 | 0.27 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
25300 |
0.1 |
| chr1_153521085_153521570 | 0.27 |
S100A3 |
S100 calcium binding protein A3 |
275 |
0.76 |
| chr11_12792556_12792707 | 0.27 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
26043 |
0.2 |
| chr1_119822332_119822483 | 0.27 |
ENSG00000238679 |
. |
20743 |
0.2 |
| chr12_52810052_52810346 | 0.27 |
RP11-1020M18.10 |
|
8835 |
0.09 |
| chr1_115873923_115874074 | 0.26 |
NGF |
nerve growth factor (beta polypeptide) |
6859 |
0.27 |
| chr15_83222576_83222727 | 0.26 |
RP11-152F13.10 |
|
2031 |
0.21 |
| chr6_33853805_33853956 | 0.26 |
MLN |
motilin |
82092 |
0.08 |
| chr9_73569352_73569503 | 0.26 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
85453 |
0.1 |
| chr17_2869063_2869214 | 0.26 |
CTD-3060P21.1 |
|
51 |
0.98 |
| chr1_39583247_39583398 | 0.25 |
ENSG00000206654 |
. |
3393 |
0.22 |
| chr10_64214734_64214920 | 0.25 |
ZNF365 |
zinc finger protein 365 |
65380 |
0.13 |
| chr1_27731813_27732238 | 0.25 |
GPR3 |
G protein-coupled receptor 3 |
12877 |
0.13 |
| chr17_1903178_1903329 | 0.25 |
CTD-2545H1.2 |
|
4345 |
0.12 |
| chr6_52282219_52282494 | 0.25 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
2750 |
0.34 |
| chr3_13555826_13555977 | 0.25 |
FBLN2 |
fibulin 2 |
17923 |
0.18 |
| chr9_130297629_130297780 | 0.24 |
FAM129B |
family with sequence similarity 129, member B |
33663 |
0.12 |
| chr9_71746394_71746545 | 0.24 |
TJP2 |
tight junction protein 2 |
10245 |
0.26 |
| chr9_137394449_137394849 | 0.24 |
RXRA |
retinoid X receptor, alpha |
96221 |
0.07 |
| chr3_50202328_50202494 | 0.24 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
5959 |
0.12 |
| chr5_131430060_131430455 | 0.24 |
ENSG00000253067 |
. |
14498 |
0.15 |
| chr1_226005147_226005298 | 0.24 |
EPHX1 |
epoxide hydrolase 1, microsomal (xenobiotic) |
7386 |
0.15 |
| chr1_17275358_17275509 | 0.24 |
CROCC |
ciliary rootlet coiled-coil, rootletin |
26183 |
0.11 |
| chr4_123752064_123752215 | 0.24 |
ENSG00000253069 |
. |
3970 |
0.22 |
| chr10_99356089_99356377 | 0.24 |
HOGA1 |
4-hydroxy-2-oxoglutarate aldolase 1 |
3326 |
0.16 |
| chr5_176898443_176898858 | 0.24 |
DBN1 |
drebrin 1 |
1221 |
0.28 |
| chr3_139020761_139020923 | 0.24 |
MRPS22 |
mitochondrial ribosomal protein S22 |
42019 |
0.13 |
| chr3_48636800_48636990 | 0.24 |
COL7A1 |
collagen, type VII, alpha 1 |
4195 |
0.11 |
| chr20_33893224_33893375 | 0.24 |
FAM83C |
family with sequence similarity 83, member C |
13095 |
0.1 |
| chr7_157197915_157198509 | 0.24 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
65697 |
0.11 |
| chr17_45353575_45353931 | 0.24 |
ENSG00000238419 |
. |
16967 |
0.13 |
| chr14_105439124_105439578 | 0.24 |
AHNAK2 |
AHNAK nucleoprotein 2 |
5343 |
0.17 |
| chr2_43268497_43268648 | 0.23 |
ENSG00000207087 |
. |
50060 |
0.18 |
| chr2_10276228_10276379 | 0.23 |
C2orf48 |
chromosome 2 open reading frame 48 |
5206 |
0.14 |
| chr15_90327580_90327731 | 0.23 |
MESP2 |
mesoderm posterior 2 homolog (mouse) |
8066 |
0.13 |
| chr11_121802656_121802807 | 0.23 |
ENSG00000252556 |
. |
72332 |
0.12 |
| chr16_4513814_4513965 | 0.23 |
NMRAL1 |
NmrA-like family domain containing 1 |
2286 |
0.2 |
| chr11_9384115_9384314 | 0.23 |
IPO7 |
importin 7 |
21955 |
0.16 |
| chr1_183261326_183261477 | 0.23 |
NMNAT2 |
nicotinamide nucleotide adenylyltransferase 2 |
12608 |
0.26 |
| chr4_25116416_25116567 | 0.23 |
SEPSECS |
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
45529 |
0.15 |
| chr1_228338191_228338342 | 0.23 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
713 |
0.51 |
| chr8_124553954_124554105 | 0.23 |
FBXO32 |
F-box protein 32 |
583 |
0.77 |
| chr3_52140801_52140952 | 0.23 |
LINC00696 |
long intergenic non-protein coding RNA 696 |
43309 |
0.09 |
| chr14_105437154_105437305 | 0.23 |
AHNAK2 |
AHNAK nucleoprotein 2 |
7465 |
0.16 |
| chr7_41743932_41744200 | 0.22 |
AC005027.3 |
|
867 |
0.51 |
| chr17_19234995_19235146 | 0.22 |
RP11-135L13.4 |
|
4370 |
0.13 |
| chr4_128544405_128544987 | 0.22 |
INTU |
inturned planar cell polarity protein |
270 |
0.95 |
| chr20_4356829_4357115 | 0.22 |
ADRA1D |
adrenoceptor alpha 1D |
127251 |
0.05 |
| chr1_227963929_227964080 | 0.22 |
SNAP47 |
synaptosomal-associated protein, 47kDa |
28240 |
0.14 |
| chr1_244088089_244088421 | 0.22 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
73874 |
0.11 |
| chr5_148532192_148532343 | 0.22 |
ABLIM3 |
actin binding LIM protein family, member 3 |
10657 |
0.19 |
| chr7_105470298_105470449 | 0.22 |
ATXN7L1 |
ataxin 7-like 1 |
46550 |
0.18 |
| chr17_27045400_27045747 | 0.22 |
RAB34 |
RAB34, member RAS oncogene family |
126 |
0.8 |
| chr22_45949911_45950062 | 0.21 |
FBLN1 |
fibulin 1 |
35323 |
0.15 |
| chr20_2218141_2218292 | 0.21 |
TGM3 |
transglutaminase 3 |
58431 |
0.12 |
| chr14_21570539_21570690 | 0.21 |
ZNF219 |
zinc finger protein 219 |
1124 |
0.27 |
| chr19_47407880_47408230 | 0.21 |
ARHGAP35 |
Rho GTPase activating protein 35 |
13878 |
0.17 |
| chr1_11851239_11851390 | 0.21 |
C1orf167 |
chromosome 1 open reading frame 167 |
11440 |
0.1 |
| chr7_44224907_44225058 | 0.21 |
GCK |
glucokinase (hexokinase 4) |
3604 |
0.16 |
| chr8_41575514_41575665 | 0.21 |
ANK1 |
ankyrin 1, erythrocytic |
11892 |
0.18 |
| chr13_114504859_114505144 | 0.21 |
GAS6-AS1 |
GAS6 antisense RNA 1 |
13602 |
0.2 |
| chr5_57537546_57537697 | 0.21 |
ENSG00000238899 |
. |
171606 |
0.03 |
| chr7_30844366_30844720 | 0.21 |
AC004691.5 |
|
1517 |
0.41 |
| chr2_216613985_216614378 | 0.21 |
ENSG00000212055 |
. |
129461 |
0.05 |
| chr17_13639928_13640079 | 0.21 |
ENSG00000236088 |
. |
55864 |
0.16 |
| chr17_16767898_16768322 | 0.21 |
AC022596.2 |
|
12888 |
0.13 |
| chr1_201504197_201504388 | 0.21 |
CSRP1 |
cysteine and glycine-rich protein 1 |
25708 |
0.14 |
| chr15_74721479_74721779 | 0.20 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
4372 |
0.17 |
| chr15_63270206_63270357 | 0.20 |
TPM1 |
tropomyosin 1 (alpha) |
64550 |
0.1 |
| chr5_136554415_136554566 | 0.20 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
94737 |
0.09 |
| chr7_51114106_51114257 | 0.20 |
RP4-724E13.2 |
|
29021 |
0.24 |
| chr17_73805763_73805914 | 0.20 |
UNK |
unkempt family zinc finger |
9982 |
0.09 |
| chr18_20171274_20171425 | 0.20 |
ENSG00000206827 |
. |
56196 |
0.15 |
| chr9_34253979_34254272 | 0.20 |
ENSG00000222426 |
. |
28543 |
0.11 |
| chr8_103802696_103803356 | 0.20 |
ENSG00000266799 |
. |
57079 |
0.11 |
| chr1_54666582_54666967 | 0.20 |
MRPL37 |
mitochondrial ribosomal protein L37 |
882 |
0.39 |
| chr12_52604840_52604991 | 0.20 |
KRT80 |
keratin 80 |
19131 |
0.1 |
| chr10_29760583_29760940 | 0.20 |
SVIL |
supervillin |
24686 |
0.2 |
| chr2_192251959_192252110 | 0.20 |
MYO1B |
myosin IB |
4826 |
0.27 |
| chr7_28076131_28076580 | 0.20 |
JAZF1 |
JAZF zinc finger 1 |
15937 |
0.26 |
| chr7_17372800_17372951 | 0.20 |
AC003075.4 |
|
33894 |
0.17 |
| chr19_33885180_33885411 | 0.19 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
20077 |
0.19 |
| chr15_29570484_29570635 | 0.19 |
NDNL2 |
necdin-like 2 |
8526 |
0.25 |
| chr16_75290840_75291057 | 0.19 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
770 |
0.56 |
| chr1_156094375_156094823 | 0.19 |
LMNA |
lamin A/C |
1352 |
0.29 |
| chr12_57542255_57543089 | 0.19 |
RP11-545N8.3 |
|
1270 |
0.29 |
| chr2_27937945_27938621 | 0.19 |
AC074091.13 |
Uncharacterized protein |
316 |
0.86 |
| chr7_137564480_137564636 | 0.19 |
DGKI |
diacylglycerol kinase, iota |
32720 |
0.18 |
| chr19_1168901_1169806 | 0.19 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
411 |
0.76 |
| chr12_3226263_3226517 | 0.19 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
32573 |
0.17 |
| chr13_51162602_51162753 | 0.19 |
DLEU7-AS1 |
DLEU7 antisense RNA 1 |
219315 |
0.02 |
| chr1_153572031_153572387 | 0.19 |
S100A16 |
S100 calcium binding protein A16 |
8589 |
0.08 |
| chr9_137516691_137516842 | 0.19 |
COL5A1 |
collagen, type V, alpha 1 |
16854 |
0.2 |
| chr2_39743971_39744122 | 0.19 |
AC007246.3 |
|
39 |
0.98 |
| chr8_124564532_124564683 | 0.19 |
FBXO32 |
F-box protein 32 |
11161 |
0.19 |
| chr22_36727795_36728637 | 0.19 |
ENSG00000266345 |
. |
15970 |
0.18 |
| chr2_237771976_237772127 | 0.19 |
ENSG00000202341 |
. |
151897 |
0.04 |
| chr1_156675699_156676565 | 0.19 |
CRABP2 |
cellular retinoic acid binding protein 2 |
524 |
0.6 |
| chr3_52039773_52040512 | 0.19 |
RPL29 |
ribosomal protein L29 |
10184 |
0.09 |
| chr2_206628931_206629328 | 0.19 |
AC007362.3 |
|
399 |
0.9 |
| chr18_20406433_20406706 | 0.19 |
RBBP8 |
retinoblastoma binding protein 8 |
28345 |
0.16 |
| chr3_25466068_25466219 | 0.19 |
RARB |
retinoic acid receptor, beta |
3611 |
0.33 |
| chr9_137222033_137222297 | 0.19 |
RXRA |
retinoid X receptor, alpha |
3739 |
0.3 |
| chr14_51909942_51910093 | 0.19 |
FRMD6 |
FERM domain containing 6 |
45838 |
0.16 |
| chr3_58192334_58192485 | 0.19 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
1859 |
0.34 |
| chr16_65606309_65606460 | 0.19 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
450116 |
0.01 |
| chr4_189048820_189048971 | 0.19 |
ENSG00000201085 |
. |
6107 |
0.18 |
| chr19_13107890_13108177 | 0.19 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
1381 |
0.23 |
| chr12_106502007_106502305 | 0.19 |
RP11-114F10.3 |
|
5215 |
0.26 |
| chr2_101728983_101729400 | 0.19 |
ENSG00000265860 |
. |
19160 |
0.19 |
| chr18_46074651_46074802 | 0.19 |
CTIF |
CBP80/20-dependent translation initiation factor |
8260 |
0.26 |
| chr6_150590777_150590928 | 0.19 |
ENSG00000201628 |
. |
57047 |
0.13 |
| chr5_52324020_52324171 | 0.19 |
CTD-2175A23.1 |
|
37987 |
0.15 |
| chr18_42834862_42835013 | 0.19 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
41977 |
0.19 |
| chr5_139031059_139031535 | 0.19 |
CXXC5 |
CXXC finger protein 5 |
723 |
0.71 |
| chr15_40628103_40628665 | 0.19 |
C15orf52 |
chromosome 15 open reading frame 52 |
1857 |
0.16 |
| chr17_42665074_42665225 | 0.19 |
FZD2 |
frizzled family receptor 2 |
30224 |
0.14 |
| chr20_49983402_49983553 | 0.19 |
ENSG00000263645 |
. |
10381 |
0.28 |
| chr4_129309482_129309633 | 0.19 |
PGRMC2 |
progesterone receptor membrane component 2 |
99573 |
0.09 |
| chr10_5594332_5594536 | 0.18 |
CALML3-AS1 |
CALML3 antisense RNA 1 |
26225 |
0.13 |
| chr9_95501833_95502258 | 0.18 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
25049 |
0.17 |
| chr5_71506479_71506741 | 0.18 |
MAP1B |
microtubule-associated protein 1B |
31155 |
0.18 |
| chr19_48970127_48970278 | 0.18 |
CTC-273B12.5 |
|
8 |
0.93 |
| chr5_14181628_14181857 | 0.18 |
TRIO |
trio Rho guanine nucleotide exchange factor |
2165 |
0.48 |
| chr15_51520595_51520807 | 0.18 |
CYP19A1 |
cytochrome P450, family 19, subfamily A, polypeptide 1 |
13280 |
0.18 |
| chr18_60544137_60544288 | 0.18 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
26171 |
0.18 |
| chr19_30154923_30155074 | 0.18 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
965 |
0.62 |
| chr7_47334154_47334412 | 0.18 |
TNS3 |
tensin 3 |
14367 |
0.31 |
| chr4_141177970_141178121 | 0.18 |
SCOC |
short coiled-coil protein |
395 |
0.88 |
| chr1_16466515_16466919 | 0.18 |
RP11-276H7.2 |
|
14989 |
0.11 |
| chr11_44613969_44614120 | 0.18 |
CD82 |
CD82 molecule |
4362 |
0.23 |
| chr15_70087017_70087168 | 0.18 |
ENSG00000215958 |
. |
9249 |
0.28 |
| chr2_238407130_238407281 | 0.18 |
MLPH |
melanophilin |
11286 |
0.19 |
| chrX_45211105_45211256 | 0.18 |
RP11-342D14.1 |
|
31477 |
0.25 |
| chr7_47560263_47560414 | 0.18 |
TNS3 |
tensin 3 |
18537 |
0.28 |
| chr22_50741961_50742432 | 0.18 |
PLXNB2 |
plexin B2 |
2305 |
0.15 |
| chr17_43205364_43205515 | 0.18 |
PLCD3 |
phospholipase C, delta 3 |
4452 |
0.12 |
| chr17_75282058_75282603 | 0.18 |
SEPT9 |
septin 9 |
1643 |
0.44 |
| chr1_245986181_245986332 | 0.18 |
RP11-522M21.3 |
|
146476 |
0.04 |
| chr1_227161852_227162199 | 0.18 |
ADCK3 |
aarF domain containing kinase 3 |
8643 |
0.21 |
| chr3_185432797_185433008 | 0.18 |
C3orf65 |
chromosome 3 open reading frame 65 |
1822 |
0.42 |
| chr14_34155007_34155158 | 0.18 |
NPAS3 |
neuronal PAS domain protein 3 |
49365 |
0.19 |
| chr1_17880750_17880901 | 0.18 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
14495 |
0.24 |
| chr3_87027489_87027640 | 0.18 |
VGLL3 |
vestigial like 3 (Drosophila) |
12288 |
0.31 |
| chr11_12136448_12136727 | 0.18 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
1656 |
0.5 |
| chr11_13140850_13141001 | 0.18 |
ENSG00000266625 |
. |
155738 |
0.04 |
| chr7_126891938_126892869 | 0.18 |
GRM8 |
glutamate receptor, metabotropic 8 |
32 |
0.98 |
| chr2_237657369_237658039 | 0.18 |
ACKR3 |
atypical chemokine receptor 3 |
179420 |
0.03 |
| chr14_103984756_103984907 | 0.18 |
CKB |
creatine kinase, brain |
2607 |
0.14 |
| chr5_54896290_54897472 | 0.17 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
66003 |
0.11 |
| chr16_73024351_73024526 | 0.17 |
ENSG00000221799 |
. |
5682 |
0.23 |
| chr2_121372137_121372546 | 0.17 |
ENSG00000201006 |
. |
36508 |
0.21 |
| chr17_75321642_75321958 | 0.17 |
SEPT9 |
septin 9 |
3108 |
0.28 |
| chr7_30839778_30839929 | 0.17 |
AC004691.5 |
|
3173 |
0.25 |
| chr5_67614932_67615223 | 0.17 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
26681 |
0.26 |
| chr1_206677916_206678151 | 0.17 |
RP11-534L20.5 |
|
752 |
0.56 |
| chr20_10055473_10055624 | 0.17 |
ENSG00000264599 |
. |
18580 |
0.2 |
| chr1_16500642_16500847 | 0.17 |
RP11-276H7.3 |
|
14983 |
0.1 |
| chr9_137537253_137537404 | 0.17 |
COL5A1 |
collagen, type V, alpha 1 |
3708 |
0.25 |
| chr16_89233752_89233942 | 0.17 |
CDH15 |
cadherin 15, type 1, M-cadherin (myotubule) |
4328 |
0.15 |
| chr17_16914497_16914878 | 0.17 |
MPRIP |
myosin phosphatase Rho interacting protein |
31172 |
0.14 |
| chr8_141600468_141600876 | 0.17 |
AGO2 |
argonaute RISC catalytic component 2 |
15315 |
0.22 |
| chr1_151283829_151283980 | 0.17 |
ENSG00000265753 |
. |
10465 |
0.09 |
| chr19_45601288_45601848 | 0.17 |
PPP1R37 |
protein phosphatase 1, regulatory subunit 37 |
5136 |
0.1 |
| chr11_100590809_100590960 | 0.17 |
CTD-2383M3.1 |
|
32198 |
0.17 |
| chr18_33888908_33889059 | 0.17 |
FHOD3 |
formin homology 2 domain containing 3 |
11184 |
0.27 |
| chr10_17103416_17103567 | 0.17 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
66345 |
0.12 |
| chr11_73042897_73043053 | 0.17 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
20365 |
0.12 |
| chr16_53127962_53128318 | 0.17 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
4932 |
0.27 |
| chr19_47128911_47129399 | 0.17 |
PTGIR |
prostaglandin I2 (prostacyclin) receptor (IP) |
780 |
0.43 |
| chr2_27238981_27239169 | 0.17 |
MAPRE3 |
microtubule-associated protein, RP/EB family, member 3 |
1408 |
0.2 |
| chr11_65257332_65257622 | 0.17 |
AP000769.1 |
Uncharacterized protein |
34749 |
0.08 |
| chr12_7144086_7144237 | 0.17 |
LPCAT3 |
lysophosphatidylcholine acyltransferase 3 |
18347 |
0.09 |
| chr9_130336547_130336733 | 0.17 |
FAM129B |
family with sequence similarity 129, member B |
4628 |
0.2 |
| chr2_224966741_224966907 | 0.17 |
SERPINE2 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
62788 |
0.13 |
| chr9_91348798_91348949 | 0.17 |
ENSG00000265873 |
. |
11947 |
0.3 |
| chr9_118395287_118395438 | 0.17 |
DEC1 |
deleted in esophageal cancer 1 |
491265 |
0.0 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.5 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.1 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.2 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0044256 | multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.0 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:2000649 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.1 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.0 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.0 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0019430 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.0 | GO:0009188 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0051350 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) |
| 0.0 | 0.1 | GO:0018200 | peptidyl-glutamic acid modification(GO:0018200) |
| 0.0 | 0.2 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.2 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.0 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |