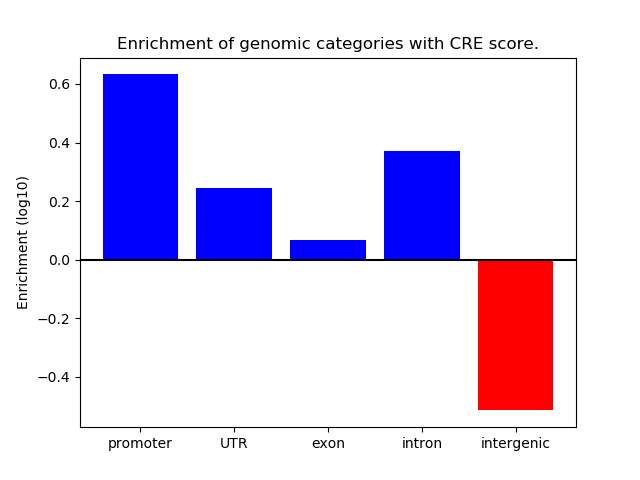
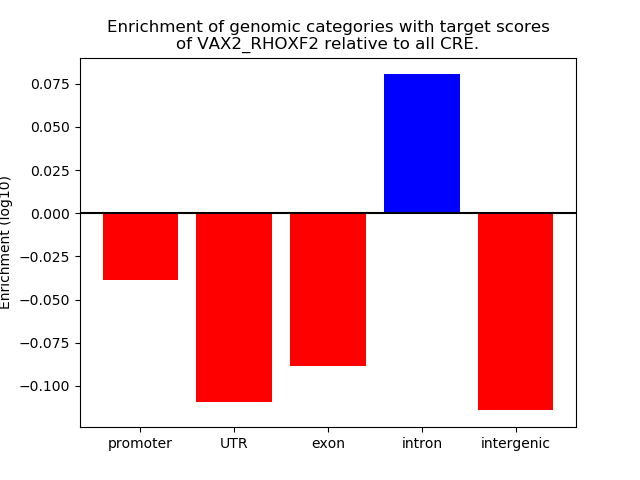
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
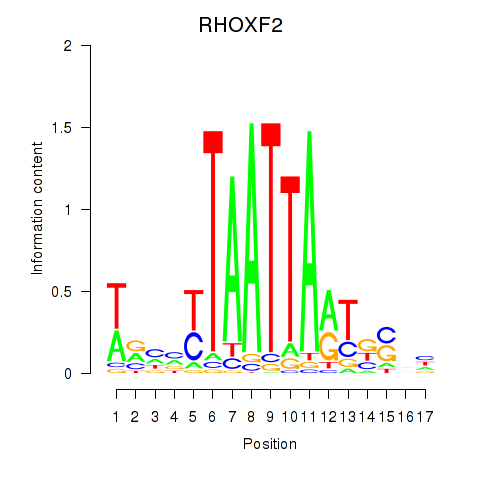
Results for VAX2_RHOXF2
Z-value: 0.51
Transcription factors associated with VAX2_RHOXF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VAX2
|
ENSG00000116035.2 | ventral anterior homeobox 2 |
|
RHOXF2
|
ENSG00000131721.4 | Rhox homeobox family member 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_119279296_119279447 | RHOXF2 | 13096 | 0.166420 | 0.72 | 3.0e-02 | Click! |
| chrX_119314962_119315113 | RHOXF2 | 22570 | 0.171382 | -0.64 | 6.4e-02 | Click! |
| chrX_119314020_119314171 | RHOXF2 | 21628 | 0.173157 | -0.63 | 6.8e-02 | Click! |
| chrX_119273447_119273598 | RHOXF2 | 18945 | 0.150548 | -0.62 | 7.7e-02 | Click! |
| chrX_119290299_119290450 | RHOXF2 | 2093 | 0.309387 | -0.20 | 6.0e-01 | Click! |
| chr2_71114393_71114674 | VAX2 | 13187 | 0.124127 | 0.73 | 2.7e-02 | Click! |
| chr2_71124315_71124466 | VAX2 | 3330 | 0.162954 | 0.61 | 7.9e-02 | Click! |
| chr2_71128195_71128714 | VAX2 | 734 | 0.548245 | 0.60 | 9.0e-02 | Click! |
| chr2_71115100_71115342 | VAX2 | 12499 | 0.124784 | 0.55 | 1.3e-01 | Click! |
| chr2_71116829_71116980 | VAX2 | 10816 | 0.126470 | 0.53 | 1.4e-01 | Click! |
Activity of the VAX2_RHOXF2 motif across conditions
Conditions sorted by the z-value of the VAX2_RHOXF2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
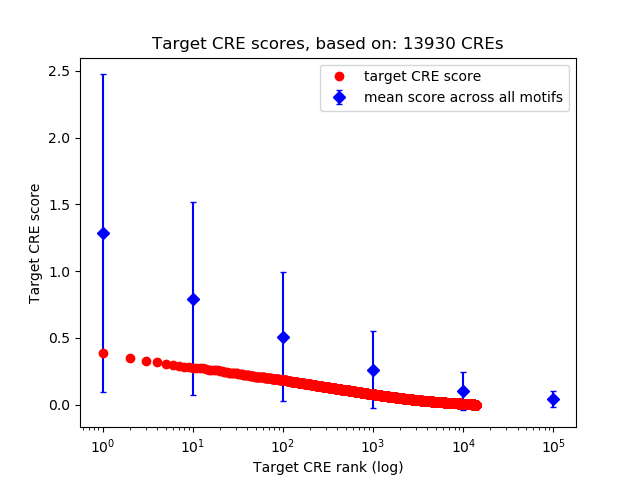
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_126941263_126941691 | 0.38 |
ENSG00000206695 |
. |
28282 |
0.26 |
| chr15_52802125_52802276 | 0.35 |
MYO5A |
myosin VA (heavy chain 12, myoxin) |
18838 |
0.2 |
| chr14_98641364_98641616 | 0.33 |
ENSG00000222066 |
. |
156597 |
0.04 |
| chr17_47825562_47826034 | 0.32 |
FAM117A |
family with sequence similarity 117, member A |
15695 |
0.14 |
| chr14_61812224_61812375 | 0.30 |
PRKCH |
protein kinase C, eta |
1257 |
0.5 |
| chrX_11784442_11784722 | 0.29 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
6835 |
0.32 |
| chr5_140934914_140935065 | 0.29 |
CTD-2024I7.13 |
|
2889 |
0.17 |
| chr10_48508940_48509161 | 0.28 |
GDF10 |
growth differentiation factor 10 |
70074 |
0.1 |
| chr1_169571123_169571274 | 0.28 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
15372 |
0.19 |
| chr19_47948776_47948927 | 0.27 |
MEIS3 |
Meis homeobox 3 |
26071 |
0.11 |
| chr14_98605614_98605900 | 0.27 |
C14orf64 |
chromosome 14 open reading frame 64 |
161296 |
0.04 |
| chr4_153000457_153000739 | 0.27 |
ENSG00000253077 |
. |
107833 |
0.07 |
| chr3_121990121_121990409 | 0.27 |
ENSG00000221474 |
. |
10435 |
0.16 |
| chr11_9416407_9416558 | 0.27 |
IPO7 |
importin 7 |
10282 |
0.16 |
| chr10_22899839_22899998 | 0.26 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
19276 |
0.27 |
| chr17_14101706_14102107 | 0.26 |
AC005224.2 |
|
11899 |
0.2 |
| chr15_60850340_60850622 | 0.26 |
CTD-2501E16.2 |
|
28309 |
0.16 |
| chr3_56963223_56963374 | 0.26 |
ARHGEF3-AS1 |
ARHGEF3 antisense RNA 1 |
10770 |
0.22 |
| chr2_43388712_43388910 | 0.26 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
64937 |
0.11 |
| chr6_158677695_158677913 | 0.25 |
TULP4 |
tubby like protein 4 |
55888 |
0.12 |
| chr5_146138797_146138948 | 0.25 |
ENSG00000265875 |
. |
24474 |
0.23 |
| chr1_175157741_175158025 | 0.25 |
KIAA0040 |
KIAA0040 |
4007 |
0.31 |
| chr2_122458404_122458555 | 0.24 |
ENSG00000238341 |
. |
5154 |
0.2 |
| chr15_85302679_85303048 | 0.24 |
RP11-7M10.2 |
|
6510 |
0.15 |
| chr6_13686993_13687323 | 0.24 |
RANBP9 |
RAN binding protein 9 |
22 |
0.98 |
| chr16_68502814_68502979 | 0.24 |
ENSG00000199263 |
. |
5671 |
0.17 |
| chr2_106545238_106545692 | 0.24 |
AC009505.2 |
|
71832 |
0.11 |
| chr17_71720142_71720293 | 0.24 |
SDK2 |
sidekick cell adhesion molecule 2 |
79989 |
0.1 |
| chr14_91858695_91858977 | 0.24 |
CCDC88C |
coiled-coil domain containing 88C |
24854 |
0.2 |
| chr15_89170331_89170558 | 0.23 |
AEN |
apoptosis enhancing nuclease |
5843 |
0.16 |
| chr11_30346022_30346188 | 0.23 |
ARL14EP |
ADP-ribosylation factor-like 14 effector protein |
1507 |
0.54 |
| chr17_14094666_14094817 | 0.23 |
ENSG00000252305 |
. |
14293 |
0.19 |
| chr4_78749172_78749323 | 0.23 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
8478 |
0.26 |
| chr2_148056205_148056470 | 0.23 |
ENSG00000238860 |
. |
25202 |
0.25 |
| chr6_2893026_2893177 | 0.23 |
SERPINB9 |
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
10413 |
0.18 |
| chr15_61011663_61011814 | 0.23 |
ENSG00000212625 |
. |
17230 |
0.2 |
| chr9_34662044_34662195 | 0.22 |
CCL27 |
chemokine (C-C motif) ligand 27 |
570 |
0.53 |
| chr13_21121097_21121248 | 0.22 |
IFT88 |
intraflagellar transport 88 homolog (Chlamydomonas) |
19413 |
0.18 |
| chr11_117877070_117877221 | 0.22 |
IL10RA |
interleukin 10 receptor, alpha |
20036 |
0.16 |
| chr10_64515756_64515907 | 0.22 |
ADO |
2-aminoethanethiol (cysteamine) dioxygenase |
48685 |
0.13 |
| chr18_32701325_32701476 | 0.22 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
79786 |
0.1 |
| chr5_75600048_75600202 | 0.22 |
RP11-466P24.6 |
|
7162 |
0.3 |
| chr8_29412378_29412770 | 0.22 |
RP4-676L2.1 |
|
201887 |
0.02 |
| chrX_118813796_118814051 | 0.21 |
SEPT6 |
septin 6 |
12869 |
0.17 |
| chr1_27156953_27157104 | 0.21 |
ZDHHC18 |
zinc finger, DHHC-type containing 18 |
1467 |
0.28 |
| chr14_99680375_99680629 | 0.21 |
AL109767.1 |
|
48783 |
0.14 |
| chr12_66712818_66712969 | 0.21 |
HELB |
helicase (DNA) B |
16568 |
0.16 |
| chr6_11357904_11358055 | 0.21 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
24553 |
0.24 |
| chr17_65449492_65449678 | 0.21 |
ENSG00000244610 |
. |
3821 |
0.17 |
| chr11_65869578_65869729 | 0.21 |
PACS1 |
phosphofurin acidic cluster sorting protein 1 |
1419 |
0.28 |
| chr1_235439552_235439703 | 0.21 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
51038 |
0.11 |
| chr5_156969848_156970007 | 0.21 |
AC106801.1 |
|
23045 |
0.14 |
| chr6_159338511_159338747 | 0.21 |
ENSG00000223191 |
. |
8974 |
0.2 |
| chr3_3204987_3205138 | 0.21 |
RP11-97C16.1 |
|
10436 |
0.17 |
| chr3_112193935_112194086 | 0.21 |
BTLA |
B and T lymphocyte associated |
24195 |
0.21 |
| chr5_82767930_82768081 | 0.21 |
VCAN |
versican |
261 |
0.95 |
| chr9_71643128_71643290 | 0.21 |
FXN |
frataxin |
6966 |
0.23 |
| chr20_43628950_43629101 | 0.21 |
ENSG00000252021 |
. |
3964 |
0.19 |
| chr5_56129477_56129628 | 0.21 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
18151 |
0.15 |
| chr5_122851393_122851548 | 0.21 |
CSNK1G3 |
casein kinase 1, gamma 3 |
3516 |
0.28 |
| chr11_44752503_44752654 | 0.21 |
TSPAN18 |
tetraspanin 18 |
3854 |
0.33 |
| chr13_48886676_48886893 | 0.21 |
RB1 |
retinoblastoma 1 |
8873 |
0.27 |
| chr3_16356148_16356299 | 0.20 |
RP11-415F23.2 |
|
277 |
0.9 |
| chr6_108114469_108114620 | 0.20 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
21089 |
0.25 |
| chr8_21823445_21823677 | 0.20 |
XPO7 |
exportin 7 |
188 |
0.94 |
| chr7_21471999_21472183 | 0.20 |
SP4 |
Sp4 transcription factor |
4430 |
0.22 |
| chr5_75688534_75688866 | 0.20 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
10374 |
0.28 |
| chr16_74573386_74573537 | 0.20 |
GLG1 |
golgi glycoprotein 1 |
23762 |
0.19 |
| chr14_68808631_68808789 | 0.20 |
RAD51B |
RAD51 paralog B |
69517 |
0.12 |
| chr10_102996848_102997121 | 0.20 |
LBX1-AS1 |
LBX1 antisense RNA 1 (head to head) |
711 |
0.61 |
| chr5_118654433_118654664 | 0.20 |
ENSG00000243333 |
. |
12222 |
0.19 |
| chr1_167435126_167435452 | 0.20 |
RP11-104L21.2 |
|
7391 |
0.22 |
| chr3_184274253_184274404 | 0.20 |
EIF2B5-AS1 |
EIF2B5 antisense RNA 1 |
378 |
0.86 |
| chr19_36000408_36000843 | 0.20 |
DMKN |
dermokine |
488 |
0.65 |
| chr2_109603249_109603400 | 0.20 |
EDAR |
ectodysplasin A receptor |
2401 |
0.41 |
| chr5_130638723_130638874 | 0.20 |
CDC42SE2 |
CDC42 small effector 2 |
39005 |
0.21 |
| chr11_128167606_128167864 | 0.20 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
207554 |
0.02 |
| chr1_115041886_115042037 | 0.20 |
ENSG00000201114 |
. |
8615 |
0.22 |
| chr1_66712472_66712670 | 0.19 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
10759 |
0.3 |
| chr4_78777778_78777929 | 0.19 |
MRPL1 |
mitochondrial ribosomal protein L1 |
5821 |
0.28 |
| chr2_205942725_205942950 | 0.19 |
PARD3B |
par-3 family cell polarity regulator beta |
532114 |
0.0 |
| chr7_128880488_128880639 | 0.19 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
15672 |
0.16 |
| chr5_112359168_112359319 | 0.19 |
DCP2 |
decapping mRNA 2 |
46764 |
0.15 |
| chrX_20048047_20048247 | 0.19 |
ENSG00000264566 |
. |
12842 |
0.17 |
| chr6_117804732_117805043 | 0.19 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
1062 |
0.49 |
| chr4_123556022_123556173 | 0.19 |
IL21 |
interleukin 21 |
13873 |
0.21 |
| chr12_838464_838718 | 0.19 |
WNK1 |
WNK lysine deficient protein kinase 1 |
23168 |
0.18 |
| chr5_156636522_156636760 | 0.19 |
CTB-4E7.1 |
|
14749 |
0.12 |
| chr1_167615092_167615243 | 0.19 |
RP3-455J7.4 |
|
15256 |
0.19 |
| chr1_100844302_100844453 | 0.19 |
ENSG00000216067 |
. |
46 |
0.98 |
| chr7_6522910_6523061 | 0.19 |
FLJ20306 |
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
63 |
0.89 |
| chr15_60865631_60865913 | 0.19 |
RORA |
RAR-related orphan receptor A |
18968 |
0.19 |
| chr3_108566035_108566186 | 0.19 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
24491 |
0.23 |
| chr16_74620197_74620348 | 0.19 |
GLG1 |
golgi glycoprotein 1 |
20720 |
0.2 |
| chr6_647016_647167 | 0.19 |
HUS1B |
HUS1 checkpoint homolog b (S. pombe) |
9872 |
0.27 |
| chr1_29490695_29490846 | 0.19 |
RP11-242O24.5 |
|
11769 |
0.16 |
| chrX_39170629_39170782 | 0.19 |
ENSG00000207122 |
. |
230167 |
0.02 |
| chr3_112040032_112040183 | 0.19 |
CD200 |
CD200 molecule |
11087 |
0.23 |
| chr7_12254768_12254919 | 0.19 |
TMEM106B |
transmembrane protein 106B |
3894 |
0.37 |
| chr12_10282940_10283100 | 0.19 |
CLEC7A |
C-type lectin domain family 7, member A |
164 |
0.93 |
| chr17_33863325_33863476 | 0.19 |
SLFN12L |
schlafen family member 12-like |
1480 |
0.27 |
| chr6_157109556_157109748 | 0.19 |
RP11-230C9.3 |
|
8063 |
0.2 |
| chr20_34234716_34234867 | 0.19 |
ENSG00000252549 |
. |
561 |
0.6 |
| chr5_171611435_171611646 | 0.19 |
STK10 |
serine/threonine kinase 10 |
3850 |
0.23 |
| chr2_15318659_15318819 | 0.18 |
NBAS |
neuroblastoma amplified sequence |
11782 |
0.32 |
| chr2_166185054_166185205 | 0.18 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
32846 |
0.22 |
| chr12_12775741_12775989 | 0.18 |
CREBL2 |
cAMP responsive element binding protein-like 2 |
11039 |
0.2 |
| chr16_4114751_4115046 | 0.18 |
RP11-462G12.4 |
|
32885 |
0.16 |
| chr9_92032244_92032395 | 0.18 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
1429 |
0.51 |
| chr10_4121743_4121894 | 0.18 |
KLF6 |
Kruppel-like factor 6 |
294345 |
0.01 |
| chr17_33348954_33349130 | 0.18 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
612 |
0.63 |
| chr6_156720023_156720174 | 0.18 |
ENSG00000212295 |
. |
20214 |
0.3 |
| chr17_40700754_40700905 | 0.18 |
HSD17B1 |
hydroxysteroid (17-beta) dehydrogenase 1 |
403 |
0.66 |
| chr18_60873699_60873977 | 0.18 |
ENSG00000238988 |
. |
11940 |
0.23 |
| chr3_184325833_184325984 | 0.18 |
EPHB3 |
EPH receptor B3 |
46336 |
0.13 |
| chr6_119226723_119226983 | 0.18 |
ASF1A |
anti-silencing function 1A histone chaperone |
11469 |
0.19 |
| chr14_52381180_52381331 | 0.18 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
1425 |
0.37 |
| chr6_122733810_122733961 | 0.18 |
HSF2 |
heat shock transcription factor 2 |
9416 |
0.25 |
| chr2_106522886_106523037 | 0.18 |
AC009505.2 |
|
49328 |
0.16 |
| chr6_112020418_112020739 | 0.18 |
FYN |
FYN oncogene related to SRC, FGR, YES |
20687 |
0.21 |
| chr6_135414750_135415028 | 0.18 |
HBS1L |
HBS1-like (S. cerevisiae) |
9305 |
0.22 |
| chr10_8130727_8131157 | 0.17 |
GATA3 |
GATA binding protein 3 |
34173 |
0.24 |
| chr14_53258584_53258855 | 0.17 |
GNPNAT1 |
glucosamine-phosphate N-acetyltransferase 1 |
333 |
0.86 |
| chr20_57722289_57722440 | 0.17 |
ZNF831 |
zinc finger protein 831 |
43711 |
0.15 |
| chr17_63031973_63032289 | 0.17 |
RP11-583F2.5 |
|
15896 |
0.16 |
| chr3_42258092_42258243 | 0.17 |
ENSG00000222872 |
. |
5165 |
0.23 |
| chr14_92973076_92973375 | 0.17 |
RIN3 |
Ras and Rab interactor 3 |
6893 |
0.29 |
| chr4_20916404_20916555 | 0.17 |
KCNIP4 |
Kv channel interacting protein 4 |
69153 |
0.13 |
| chr3_32998308_32998459 | 0.17 |
CCR4 |
chemokine (C-C motif) receptor 4 |
5317 |
0.28 |
| chr16_11346729_11346935 | 0.17 |
SOCS1 |
suppressor of cytokine signaling 1 |
3204 |
0.12 |
| chr10_16877098_16877261 | 0.17 |
RSU1 |
Ras suppressor protein 1 |
17652 |
0.26 |
| chr2_30557961_30558163 | 0.17 |
ENSG00000221377 |
. |
97252 |
0.07 |
| chr5_49991075_49991374 | 0.17 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
27833 |
0.27 |
| chrY_7194717_7195015 | 0.17 |
ENSG00000252633 |
. |
2230 |
0.31 |
| chr7_25004547_25004698 | 0.17 |
OSBPL3 |
oxysterol binding protein-like 3 |
15138 |
0.26 |
| chr5_64919668_64919819 | 0.17 |
TRIM23 |
tripartite motif containing 23 |
380 |
0.59 |
| chr15_93372987_93373270 | 0.17 |
FAM174B |
family with sequence similarity 174, member B |
20100 |
0.15 |
| chr10_30991700_30991887 | 0.17 |
SVILP1 |
supervillin pseudogene 1 |
6943 |
0.27 |
| chr5_114715684_114715835 | 0.17 |
CCDC112 |
coiled-coil domain containing 112 |
83231 |
0.09 |
| chr4_109022417_109022568 | 0.17 |
LEF1 |
lymphoid enhancer-binding factor 1 |
64965 |
0.11 |
| chr2_158768231_158768382 | 0.17 |
ENSG00000251980 |
. |
978 |
0.56 |
| chr11_65107055_65107206 | 0.17 |
DPF2 |
D4, zinc and double PHD fingers family 2 |
5792 |
0.12 |
| chr14_22535112_22535263 | 0.17 |
ENSG00000238634 |
. |
75700 |
0.11 |
| chr3_14467854_14468179 | 0.17 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
6162 |
0.25 |
| chr13_42969980_42970150 | 0.17 |
AKAP11 |
A kinase (PRKA) anchor protein 11 |
123776 |
0.06 |
| chr2_235395214_235395372 | 0.17 |
ARL4C |
ADP-ribosylation factor-like 4C |
9951 |
0.32 |
| chr7_138768806_138768957 | 0.17 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
4868 |
0.24 |
| chr5_55443907_55444286 | 0.17 |
ENSG00000223003 |
. |
1423 |
0.42 |
| chr8_121714189_121714356 | 0.17 |
RP11-713M15.1 |
|
59221 |
0.14 |
| chr21_43810236_43810387 | 0.17 |
TMPRSS3 |
transmembrane protease, serine 3 |
5741 |
0.14 |
| chr10_6087007_6087203 | 0.17 |
IL2RA |
interleukin 2 receptor, alpha |
17148 |
0.13 |
| chr22_27066729_27066953 | 0.17 |
CRYBA4 |
crystallin, beta A4 |
48913 |
0.13 |
| chr16_3865090_3865241 | 0.16 |
CREBBP |
CREB binding protein |
34297 |
0.17 |
| chr5_118678786_118679078 | 0.16 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
10062 |
0.21 |
| chr8_81976580_81976731 | 0.16 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
47648 |
0.18 |
| chr22_40340974_40341233 | 0.16 |
GRAP2 |
GRB2-related adaptor protein 2 |
1718 |
0.34 |
| chr6_133057981_133058132 | 0.16 |
VNN3 |
vanin 3 |
2152 |
0.22 |
| chr2_63287841_63287992 | 0.16 |
OTX1 |
orthodenticle homeobox 1 |
9979 |
0.23 |
| chr10_102989164_102989315 | 0.16 |
LBX1-AS1 |
LBX1 antisense RNA 1 (head to head) |
112 |
0.75 |
| chr17_8311463_8311614 | 0.16 |
NDEL1 |
nudE neurodevelopment protein 1-like 1 |
4911 |
0.12 |
| chr6_35649971_35650157 | 0.16 |
FKBP5 |
FK506 binding protein 5 |
6628 |
0.14 |
| chr1_155910079_155910230 | 0.16 |
RXFP4 |
relaxin/insulin-like family peptide receptor 4 |
1326 |
0.24 |
| chr11_35020496_35020647 | 0.16 |
PDHX |
pyruvate dehydrogenase complex, component X |
21240 |
0.2 |
| chr8_126930029_126930180 | 0.16 |
ENSG00000206695 |
. |
16909 |
0.3 |
| chr3_53072608_53072759 | 0.16 |
SFMBT1 |
Scm-like with four mbt domains 1 |
6598 |
0.19 |
| chr12_58147924_58148156 | 0.16 |
MARCH9 |
membrane-associated ring finger (C3HC4) 9 |
841 |
0.29 |
| chr18_67554931_67555082 | 0.16 |
CD226 |
CD226 molecule |
59649 |
0.15 |
| chr4_57687212_57687509 | 0.16 |
SPINK2 |
serine peptidase inhibitor, Kazal type 2 (acrosin-trypsin inhibitor) |
500 |
0.74 |
| chr8_27223358_27224128 | 0.16 |
PTK2B |
protein tyrosine kinase 2 beta |
14425 |
0.22 |
| chr17_73374792_73374943 | 0.16 |
RP11-16C1.2 |
|
5050 |
0.11 |
| chr6_149404447_149404642 | 0.16 |
RP11-162J8.3 |
|
50835 |
0.15 |
| chr19_12442701_12443014 | 0.16 |
ZNF563 |
zinc finger protein 563 |
1449 |
0.33 |
| chr6_44797257_44797514 | 0.16 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
125862 |
0.06 |
| chr8_127567681_127568033 | 0.16 |
RP11-103H7.5 |
|
1284 |
0.38 |
| chr13_41574006_41574157 | 0.16 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
17663 |
0.19 |
| chr9_86303237_86303482 | 0.16 |
RP11-522I20.3 |
|
19150 |
0.17 |
| chr1_226911013_226911201 | 0.16 |
ITPKB |
inositol-trisphosphate 3-kinase B |
14052 |
0.21 |
| chr18_19178052_19178356 | 0.16 |
ESCO1 |
establishment of sister chromatid cohesion N-acetyltransferase 1 |
2493 |
0.23 |
| chr12_111005224_111005381 | 0.16 |
PPTC7 |
PTC7 protein phosphatase homolog (S. cerevisiae) |
15823 |
0.14 |
| chr1_235014905_235015056 | 0.16 |
ENSG00000239690 |
. |
24953 |
0.21 |
| chr16_73092870_73093114 | 0.16 |
ZFHX3 |
zinc finger homeobox 3 |
605 |
0.79 |
| chr4_38518191_38518342 | 0.16 |
RP11-617D20.1 |
|
107930 |
0.07 |
| chr14_92322968_92323119 | 0.16 |
TC2N |
tandem C2 domains, nuclear |
10830 |
0.2 |
| chr3_101229742_101229893 | 0.16 |
SENP7 |
SUMO1/sentrin specific peptidase 7 |
2211 |
0.26 |
| chr2_106375349_106375537 | 0.16 |
NCK2 |
NCK adaptor protein 2 |
13255 |
0.28 |
| chr2_222385882_222386088 | 0.16 |
ENSG00000221432 |
. |
1608 |
0.44 |
| chr9_31336901_31337115 | 0.16 |
ENSG00000211510 |
. |
562782 |
0.0 |
| chr5_75626248_75626505 | 0.16 |
RP11-466P24.6 |
|
19089 |
0.25 |
| chrX_78338727_78338878 | 0.15 |
GPR174 |
G protein-coupled receptor 174 |
87667 |
0.1 |
| chr10_4178314_4178722 | 0.15 |
KLF6 |
Kruppel-like factor 6 |
351045 |
0.01 |
| chr5_141503692_141503877 | 0.15 |
NDFIP1 |
Nedd4 family interacting protein 1 |
15714 |
0.23 |
| chrX_20456293_20456479 | 0.15 |
ENSG00000252978 |
. |
13840 |
0.3 |
| chr13_48760783_48760934 | 0.15 |
ITM2B |
integral membrane protein 2B |
46436 |
0.16 |
| chr5_56145818_56146186 | 0.15 |
ENSG00000238717 |
. |
6973 |
0.16 |
| chr15_38959465_38959802 | 0.15 |
C15orf53 |
chromosome 15 open reading frame 53 |
29166 |
0.22 |
| chr6_35635066_35635406 | 0.15 |
ENSG00000265527 |
. |
2670 |
0.2 |
| chr19_14461542_14461693 | 0.15 |
CD97 |
CD97 molecule |
30351 |
0.13 |
| chr2_106402335_106402603 | 0.15 |
NCK2 |
NCK adaptor protein 2 |
30546 |
0.22 |
| chr5_61873952_61874158 | 0.15 |
LRRC70 |
leucine rich repeat containing 70 |
507 |
0.52 |
| chr17_4263545_4263821 | 0.15 |
ENSG00000263882 |
. |
4065 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:1904035 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.0 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0060510 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0050856 | regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.0 | 0.0 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:0097300 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.0 | GO:0033088 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.2 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:2001234 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1904376 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.1 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.0 | GO:1903312 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.1 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 | 0.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.3 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.0 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0033081 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.0 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.0 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.0 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.3 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.2 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.0 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.0 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.8 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.0 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.0 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |