Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
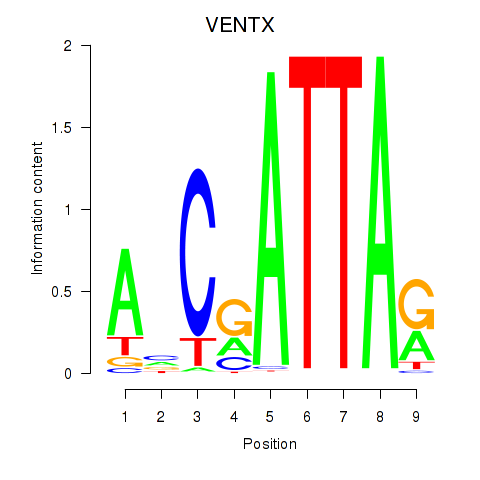
Results for VENTX
Z-value: 1.03
Transcription factors associated with VENTX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VENTX
|
ENSG00000151650.7 | VENT homeobox |
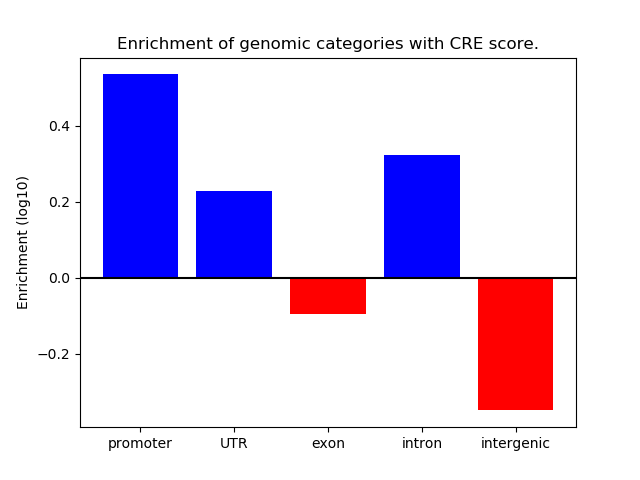
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_135051321_135051472 | VENTX | 488 | 0.691496 | 0.57 | 1.1e-01 | Click! |
| chr10_135053713_135053864 | VENTX | 2880 | 0.157198 | 0.56 | 1.2e-01 | Click! |
| chr10_135052910_135053061 | VENTX | 2077 | 0.198820 | 0.39 | 3.0e-01 | Click! |
| chr10_135048749_135049130 | VENTX | 1969 | 0.210239 | 0.35 | 3.6e-01 | Click! |
| chr10_135049859_135050010 | VENTX | 974 | 0.410139 | -0.33 | 3.8e-01 | Click! |
Activity of the VENTX motif across conditions
Conditions sorted by the z-value of the VENTX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
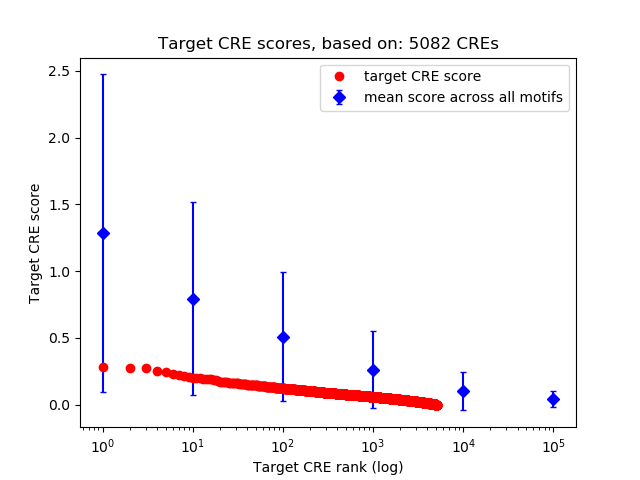
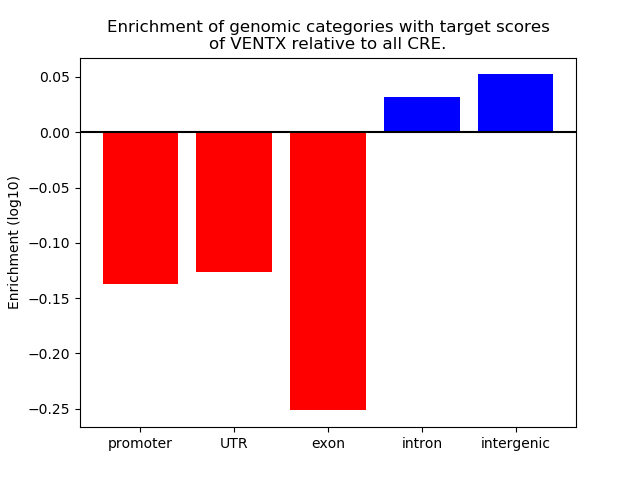
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_234755566_234755985 | 0.28 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
10504 |
0.19 |
| chr12_32268115_32268884 | 0.28 |
RP11-843B15.2 |
|
8037 |
0.23 |
| chr18_21418281_21418637 | 0.27 |
LAMA3 |
laminin, alpha 3 |
4542 |
0.28 |
| chr9_128521593_128522112 | 0.25 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
11374 |
0.27 |
| chr9_79268514_79268818 | 0.24 |
PRUNE2 |
prune homolog 2 (Drosophila) |
1201 |
0.53 |
| chr11_10661711_10661903 | 0.23 |
MRVI1 |
murine retrovirus integration site 1 homolog |
11925 |
0.19 |
| chr7_102532238_102532538 | 0.23 |
LRRC17 |
leucine rich repeat containing 17 |
21050 |
0.16 |
| chr17_78570092_78570495 | 0.22 |
RPTOR |
regulatory associated protein of MTOR, complex 1 |
51025 |
0.14 |
| chr21_18864726_18865102 | 0.21 |
ENSG00000266195 |
. |
13857 |
0.15 |
| chr4_174411560_174411794 | 0.20 |
HAND2-AS1 |
HAND2 antisense RNA 1 (head to head) |
36744 |
0.14 |
| chr3_126450507_126450658 | 0.20 |
CHCHD6 |
coiled-coil-helix-coiled-coil-helix domain containing 6 |
1175 |
0.59 |
| chr17_45302812_45302963 | 0.20 |
ENSG00000252088 |
. |
4879 |
0.14 |
| chr1_15485106_15485305 | 0.20 |
TMEM51 |
transmembrane protein 51 |
4920 |
0.24 |
| chr3_9939344_9939569 | 0.19 |
IL17RE |
interleukin 17 receptor E |
4847 |
0.09 |
| chr5_59090911_59091107 | 0.19 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
26538 |
0.25 |
| chr2_25479868_25480019 | 0.19 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
4763 |
0.25 |
| chr16_69510453_69510859 | 0.19 |
CYB5B |
cytochrome b5 type B (outer mitochondrial membrane) |
51970 |
0.1 |
| chr13_108520309_108520631 | 0.19 |
FAM155A |
family with sequence similarity 155, member A |
1387 |
0.54 |
| chr15_95389413_95389564 | 0.18 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
490285 |
0.01 |
| chr14_55369760_55370261 | 0.17 |
GCH1 |
GTP cyclohydrolase 1 |
440 |
0.84 |
| chr5_66001075_66001226 | 0.17 |
MAST4-IT1 |
MAST4 intronic transcript 1 (non-protein coding) |
42991 |
0.18 |
| chr1_200201570_200201721 | 0.17 |
ENSG00000221403 |
. |
87683 |
0.09 |
| chr12_89339824_89340184 | 0.17 |
ENSG00000238302 |
. |
336058 |
0.01 |
| chr5_139039967_139040260 | 0.17 |
CXXC5 |
CXXC finger protein 5 |
973 |
0.6 |
| chr1_240406130_240406281 | 0.17 |
FMN2 |
formin 2 |
2355 |
0.37 |
| chr9_107630817_107631231 | 0.17 |
RP11-217B7.2 |
|
58810 |
0.12 |
| chr2_46169357_46169508 | 0.16 |
PRKCE |
protein kinase C, epsilon |
58609 |
0.15 |
| chr3_57657383_57657934 | 0.16 |
DENND6A |
DENN/MADD domain containing 6A |
11105 |
0.15 |
| chr3_193716358_193716509 | 0.16 |
RP11-135A1.2 |
|
132557 |
0.05 |
| chr6_100059341_100059492 | 0.16 |
PRDM13 |
PR domain containing 13 |
4810 |
0.2 |
| chr13_98506022_98506173 | 0.16 |
ENSG00000238407 |
. |
31966 |
0.23 |
| chr3_179435073_179435364 | 0.16 |
USP13 |
ubiquitin specific peptidase 13 (isopeptidase T-3) |
4134 |
0.27 |
| chr5_139047754_139047905 | 0.16 |
CXXC5 |
CXXC finger protein 5 |
7192 |
0.22 |
| chr10_104306908_104307119 | 0.16 |
ENSG00000242311 |
. |
21291 |
0.12 |
| chr3_124352300_124352451 | 0.16 |
KALRN |
kalirin, RhoGEF kinase |
3569 |
0.28 |
| chr8_6418941_6419092 | 0.16 |
ANGPT2 |
angiopoietin 2 |
1549 |
0.51 |
| chr18_74188097_74188669 | 0.16 |
ZNF516 |
zinc finger protein 516 |
14352 |
0.17 |
| chr18_59659781_59660176 | 0.15 |
RNF152 |
ring finger protein 152 |
98514 |
0.08 |
| chr13_114995824_114995975 | 0.15 |
CDC16 |
cell division cycle 16 |
4463 |
0.2 |
| chr12_66356609_66356760 | 0.15 |
RP11-366L20.3 |
|
14425 |
0.2 |
| chr18_42871797_42872157 | 0.15 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
79017 |
0.1 |
| chr2_109209852_109210198 | 0.15 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
5103 |
0.27 |
| chr2_167229684_167229835 | 0.15 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
2738 |
0.32 |
| chr4_84482718_84482982 | 0.15 |
AGPAT9 |
1-acylglycerol-3-phosphate O-acyltransferase 9 |
25286 |
0.2 |
| chr2_9142284_9142595 | 0.15 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
1355 |
0.52 |
| chr4_22515546_22516022 | 0.15 |
GPR125 |
G protein-coupled receptor 125 |
1836 |
0.52 |
| chr1_24192279_24192516 | 0.15 |
FUCA1 |
fucosidase, alpha-L- 1, tissue |
2387 |
0.17 |
| chr10_14823935_14824305 | 0.15 |
FAM107B |
family with sequence similarity 107, member B |
7224 |
0.22 |
| chr3_127606245_127606396 | 0.15 |
KBTBD12 |
kelch repeat and BTB (POZ) domain containing 12 |
27755 |
0.19 |
| chr12_90644496_90644653 | 0.15 |
ENSG00000252823 |
. |
496738 |
0.0 |
| chr10_101280980_101281131 | 0.14 |
RP11-129J12.2 |
|
6145 |
0.2 |
| chr11_102321739_102322277 | 0.14 |
TMEM123 |
transmembrane protein 123 |
46 |
0.96 |
| chr11_1930227_1930378 | 0.14 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
10490 |
0.1 |
| chr10_126846033_126846605 | 0.14 |
CTBP2 |
C-terminal binding protein 2 |
966 |
0.69 |
| chr18_59615578_59615729 | 0.14 |
RNF152 |
ring finger protein 152 |
54189 |
0.17 |
| chr11_130671452_130671603 | 0.14 |
SNX19 |
sorting nexin 19 |
92242 |
0.09 |
| chr18_9016875_9017135 | 0.14 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
85623 |
0.08 |
| chr8_30358272_30358497 | 0.14 |
RBPMS |
RNA binding protein with multiple splicing |
58251 |
0.13 |
| chr7_115994446_115994930 | 0.14 |
ENSG00000216076 |
. |
8690 |
0.21 |
| chr10_104470200_104470393 | 0.14 |
ARL3 |
ADP-ribosylation factor-like 3 |
3868 |
0.18 |
| chr3_71450784_71450957 | 0.14 |
FOXP1 |
forkhead box P1 |
96959 |
0.08 |
| chr3_123476973_123477366 | 0.14 |
MYLK |
myosin light chain kinase |
35519 |
0.15 |
| chr12_92540486_92540987 | 0.14 |
RP11-796E2.4 |
|
779 |
0.47 |
| chr8_101503147_101503298 | 0.14 |
KB-1615E4.3 |
|
1534 |
0.38 |
| chr1_55038235_55038416 | 0.14 |
ACOT11 |
acyl-CoA thioesterase 11 |
24424 |
0.14 |
| chr5_82293579_82293730 | 0.14 |
ENSG00000238835 |
. |
66502 |
0.12 |
| chr10_130826174_130826462 | 0.14 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
439130 |
0.01 |
| chr5_13591669_13591820 | 0.14 |
CTB-51A17.1 |
|
268703 |
0.02 |
| chr10_127789954_127790105 | 0.14 |
ENSG00000222740 |
. |
44122 |
0.17 |
| chr3_150831573_150831724 | 0.13 |
MED12L |
mediator complex subunit 12-like |
26967 |
0.18 |
| chr3_172035418_172035569 | 0.13 |
AC092964.1 |
Uncharacterized protein |
1275 |
0.51 |
| chr6_108681173_108681324 | 0.13 |
ENSG00000200799 |
. |
32029 |
0.16 |
| chr1_235242449_235242943 | 0.13 |
ENSG00000207181 |
. |
48556 |
0.13 |
| chr3_100118211_100119246 | 0.13 |
LNP1 |
leukemia NUP98 fusion partner 1 |
1309 |
0.37 |
| chr6_144203576_144203727 | 0.13 |
ZC2HC1B |
zinc finger, C2HC-type containing 1B |
1790 |
0.43 |
| chr17_61509468_61509619 | 0.13 |
RP11-269G24.3 |
|
1093 |
0.39 |
| chr5_153667998_153668149 | 0.13 |
ENSG00000221070 |
. |
51613 |
0.12 |
| chr3_153007293_153007770 | 0.13 |
ENSG00000265813 |
. |
89725 |
0.09 |
| chr1_66428861_66429012 | 0.13 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
29169 |
0.23 |
| chr3_159622701_159622989 | 0.13 |
SCHIP1 |
schwannomin interacting protein 1 |
52117 |
0.12 |
| chr12_18498312_18498592 | 0.13 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
63474 |
0.15 |
| chr1_17513258_17513499 | 0.13 |
PADI1 |
peptidyl arginine deiminase, type I |
18243 |
0.16 |
| chr12_19344339_19344496 | 0.13 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
13779 |
0.2 |
| chr11_94503265_94503531 | 0.13 |
AMOTL1 |
angiomotin like 1 |
1861 |
0.4 |
| chr3_88667276_88667427 | 0.13 |
ENSG00000207316 |
. |
187010 |
0.03 |
| chr14_60097720_60097871 | 0.13 |
RTN1 |
reticulon 1 |
263 |
0.93 |
| chr3_128914264_128914631 | 0.13 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
11682 |
0.14 |
| chr1_67397953_67399117 | 0.13 |
MIER1 |
mesoderm induction early response 1, transcriptional regulator |
2609 |
0.29 |
| chr18_20714051_20714237 | 0.13 |
CABLES1 |
Cdk5 and Abl enzyme substrate 1 |
384 |
0.85 |
| chr4_187765207_187765358 | 0.13 |
ENSG00000252382 |
. |
13328 |
0.29 |
| chr10_6289225_6289376 | 0.13 |
ENSG00000238366 |
. |
1506 |
0.4 |
| chr6_7697637_7697796 | 0.13 |
BMP6 |
bone morphogenetic protein 6 |
29314 |
0.23 |
| chr17_26497742_26497961 | 0.12 |
PYY2 |
peptide YY, 2 (pseudogene) |
56481 |
0.09 |
| chr13_72401462_72401613 | 0.12 |
DACH1 |
dachshund homolog 1 (Drosophila) |
39370 |
0.23 |
| chr15_82422353_82422648 | 0.12 |
RP11-597K23.2 |
Uncharacterized protein |
41468 |
0.14 |
| chr16_81552118_81552288 | 0.12 |
CMIP |
c-Maf inducing protein |
23249 |
0.25 |
| chr2_16195675_16195826 | 0.12 |
ENSG00000243541 |
. |
104647 |
0.07 |
| chr2_145266955_145267602 | 0.12 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
7837 |
0.25 |
| chr1_117546989_117547140 | 0.12 |
CD101 |
CD101 molecule |
2630 |
0.25 |
| chr10_62332447_62332814 | 0.12 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
209 |
0.97 |
| chr9_125766060_125766211 | 0.12 |
RABGAP1 |
RAB GTPase activating protein 1 |
29653 |
0.12 |
| chr5_67829635_67829905 | 0.12 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
241374 |
0.02 |
| chr17_14736221_14736372 | 0.12 |
ENSG00000238806 |
. |
404904 |
0.01 |
| chr6_74432714_74432936 | 0.12 |
RP11-553A21.3 |
|
26971 |
0.17 |
| chr13_111569474_111569747 | 0.12 |
ANKRD10 |
ankyrin repeat domain 10 |
2194 |
0.33 |
| chr1_246170055_246170206 | 0.12 |
RP11-83A16.1 |
|
26723 |
0.24 |
| chr5_43433106_43433363 | 0.12 |
CCL28 |
chemokine (C-C motif) ligand 28 |
20741 |
0.16 |
| chr21_16433100_16433921 | 0.12 |
NRIP1 |
nuclear receptor interacting protein 1 |
3616 |
0.34 |
| chr5_163821675_163821826 | 0.12 |
CTC-340A15.2 |
|
40591 |
0.23 |
| chr17_8525673_8525824 | 0.12 |
MYH10 |
myosin, heavy chain 10, non-muscle |
8287 |
0.22 |
| chr12_96506632_96506930 | 0.12 |
ENSG00000266889 |
. |
10754 |
0.2 |
| chr4_76491442_76491625 | 0.12 |
C4orf26 |
chromosome 4 open reading frame 26 |
10258 |
0.2 |
| chr3_189699267_189699457 | 0.12 |
ENSG00000265045 |
. |
132361 |
0.05 |
| chr7_33894654_33894805 | 0.12 |
BMPER |
BMP binding endothelial regulator |
49794 |
0.18 |
| chr8_101802288_101802439 | 0.12 |
ENSG00000202001 |
. |
34940 |
0.12 |
| chr10_4541320_4541471 | 0.12 |
ENSG00000207124 |
. |
15749 |
0.3 |
| chr5_123880441_123880592 | 0.12 |
RP11-436H11.2 |
|
184008 |
0.03 |
| chr11_82781868_82782128 | 0.12 |
RAB30 |
RAB30, member RAS oncogene family |
872 |
0.59 |
| chr18_24237652_24237890 | 0.12 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
406 |
0.83 |
| chr14_68408062_68408307 | 0.12 |
RAD51B |
RAD51 paralog B |
117923 |
0.05 |
| chr18_526772_527073 | 0.12 |
COLEC12 |
collectin sub-family member 12 |
26200 |
0.14 |
| chr7_142136472_142136889 | 0.12 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
147071 |
0.04 |
| chr17_53804938_53805089 | 0.12 |
TMEM100 |
transmembrane protein 100 |
4469 |
0.29 |
| chr7_98737042_98737396 | 0.12 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
4423 |
0.27 |
| chr5_19638971_19639122 | 0.12 |
ENSG00000264928 |
. |
96624 |
0.09 |
| chr5_79031414_79031681 | 0.12 |
CMYA5 |
cardiomyopathy associated 5 |
45847 |
0.16 |
| chr5_166406041_166406192 | 0.12 |
TENM2 |
teneurin transmembrane protein 2 |
305688 |
0.01 |
| chr5_141513836_141514000 | 0.12 |
NDFIP1 |
Nedd4 family interacting protein 1 |
25848 |
0.21 |
| chr1_180762260_180762411 | 0.12 |
ENSG00000272292 |
. |
34477 |
0.17 |
| chr2_165698921_165699158 | 0.12 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
355 |
0.83 |
| chr17_37083356_37083618 | 0.12 |
RP1-56K13.3 |
|
5465 |
0.13 |
| chr7_92364918_92365200 | 0.12 |
ENSG00000206763 |
. |
33931 |
0.19 |
| chr11_12791193_12791390 | 0.11 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
24703 |
0.2 |
| chr3_112323833_112324096 | 0.11 |
CCDC80 |
coiled-coil domain containing 80 |
5163 |
0.24 |
| chr8_37659341_37659694 | 0.11 |
GPR124 |
G protein-coupled receptor 124 |
4743 |
0.17 |
| chr2_109208546_109208697 | 0.11 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
3699 |
0.29 |
| chr2_85992466_85992617 | 0.11 |
ATOH8 |
atonal homolog 8 (Drosophila) |
11524 |
0.15 |
| chr5_173958937_173959224 | 0.11 |
MSX2 |
msh homeobox 2 |
192456 |
0.03 |
| chr17_32571205_32571405 | 0.11 |
CCL2 |
chemokine (C-C motif) ligand 2 |
10999 |
0.13 |
| chr3_159744998_159745380 | 0.11 |
LINC01100 |
long intergenic non-protein coding RNA 1100 |
11378 |
0.19 |
| chr4_74527594_74528063 | 0.11 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
41480 |
0.17 |
| chr8_81721081_81721232 | 0.11 |
ENSG00000243951 |
. |
31959 |
0.19 |
| chr9_2157005_2157156 | 0.11 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
600 |
0.78 |
| chr6_118873240_118873391 | 0.11 |
PLN |
phospholamban |
3854 |
0.36 |
| chr11_27924831_27924982 | 0.11 |
RP11-587D21.4 |
|
25711 |
0.25 |
| chr1_205711446_205711715 | 0.11 |
NUCKS1 |
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
7824 |
0.14 |
| chr19_16295431_16295668 | 0.11 |
FAM32A |
family with sequence similarity 32, member A |
642 |
0.59 |
| chr1_109934550_109935130 | 0.11 |
SORT1 |
sortilin 1 |
1139 |
0.45 |
| chr5_440509_440660 | 0.11 |
C5orf55 |
chromosome 5 open reading frame 55 |
2674 |
0.18 |
| chr11_34321844_34322125 | 0.11 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
56818 |
0.14 |
| chr4_140670810_140670961 | 0.11 |
ENSG00000252233 |
. |
44702 |
0.15 |
| chr11_122311887_122312369 | 0.11 |
ENSG00000252776 |
. |
7790 |
0.23 |
| chr6_140893147_140893298 | 0.11 |
ENSG00000221336 |
. |
88628 |
0.1 |
| chr17_40230079_40230230 | 0.11 |
ZNF385C |
zinc finger protein 385C |
15079 |
0.1 |
| chr7_6431958_6432109 | 0.11 |
RAC1 |
ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
17863 |
0.16 |
| chr8_117334188_117334602 | 0.11 |
ENSG00000264815 |
. |
34807 |
0.24 |
| chr9_125836491_125836671 | 0.11 |
MIR600HG |
MIR600 host gene (non-protein coding) |
36863 |
0.13 |
| chr7_29911814_29912100 | 0.11 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
37616 |
0.15 |
| chr4_146467395_146467619 | 0.11 |
SMAD1-AS1 |
SMAD1 antisense RNA 1 |
29237 |
0.17 |
| chr11_10322719_10322870 | 0.11 |
RP11-351I24.1 |
|
2457 |
0.22 |
| chr4_173734392_173734543 | 0.11 |
ENSG00000241652 |
. |
328327 |
0.01 |
| chr4_90819954_90820271 | 0.11 |
MMRN1 |
multimerin 1 |
3022 |
0.34 |
| chr6_22877485_22877772 | 0.11 |
ENSG00000207394 |
. |
247687 |
0.02 |
| chr17_34956793_34956944 | 0.11 |
MRM1 |
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
1133 |
0.4 |
| chr6_16903564_16904059 | 0.11 |
RP1-151F17.1 |
|
141668 |
0.05 |
| chr4_81257263_81257875 | 0.11 |
C4orf22 |
chromosome 4 open reading frame 22 |
646 |
0.81 |
| chr17_48501294_48501445 | 0.11 |
ACSF2 |
acyl-CoA synthetase family member 2 |
2150 |
0.18 |
| chr13_77343543_77343694 | 0.11 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
116907 |
0.06 |
| chr10_64892562_64892759 | 0.11 |
NRBF2 |
nuclear receptor binding factor 2 |
390 |
0.86 |
| chr7_81405573_81405834 | 0.11 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
5949 |
0.33 |
| chr1_15587814_15588004 | 0.11 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
9628 |
0.21 |
| chr12_46824110_46824422 | 0.11 |
SLC38A2 |
solute carrier family 38, member 2 |
57616 |
0.14 |
| chr4_55685762_55685928 | 0.11 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
161760 |
0.04 |
| chr15_100017314_100017723 | 0.11 |
MEF2A |
myocyte enhancer factor 2A |
148 |
0.96 |
| chr6_100795466_100795617 | 0.11 |
RP1-121G13.2 |
|
79453 |
0.11 |
| chr20_5868737_5868888 | 0.11 |
ENSG00000202380 |
. |
1889 |
0.33 |
| chr12_66942372_66942523 | 0.11 |
GRIP1 |
glutamate receptor interacting protein 1 |
9559 |
0.31 |
| chr11_12716200_12716351 | 0.11 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
19225 |
0.26 |
| chr17_35567851_35568148 | 0.11 |
ACACA |
acetyl-CoA carboxylase alpha |
5273 |
0.28 |
| chr10_91405210_91405876 | 0.11 |
PANK1 |
pantothenate kinase 1 |
328 |
0.49 |
| chr10_53620310_53620501 | 0.11 |
CSTF2T |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
161050 |
0.04 |
| chr3_18127970_18128121 | 0.11 |
RP11-158G18.1 |
|
323082 |
0.01 |
| chr4_36315284_36315467 | 0.11 |
DTHD1 |
death domain containing 1 |
29731 |
0.19 |
| chr6_111927188_111927462 | 0.11 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
124 |
0.96 |
| chr2_36603144_36603295 | 0.11 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
19605 |
0.27 |
| chr7_1462493_1462644 | 0.11 |
MICALL2 |
MICAL-like 2 |
36394 |
0.13 |
| chr3_42114121_42114383 | 0.10 |
TRAK1 |
trafficking protein, kinesin binding 1 |
18310 |
0.25 |
| chr16_30344348_30345176 | 0.10 |
RP11-347C12.2 |
|
1613 |
0.21 |
| chr21_27444478_27444629 | 0.10 |
APP |
amyloid beta (A4) precursor protein |
17824 |
0.24 |
| chr1_113067430_113067581 | 0.10 |
RP4-671G15.2 |
|
6442 |
0.19 |
| chr21_32574317_32574611 | 0.10 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
71925 |
0.12 |
| chr9_36101446_36101597 | 0.10 |
GLIPR2 |
GLI pathogenesis-related 2 |
35211 |
0.14 |
| chr5_134726722_134726976 | 0.10 |
H2AFY |
H2A histone family, member Y |
8052 |
0.17 |
| chr10_126433419_126434017 | 0.10 |
FAM53B |
family with sequence similarity 53, member B |
880 |
0.56 |
| chr10_115021354_115021505 | 0.10 |
ENSG00000238380 |
. |
91755 |
0.09 |
| chr3_112530371_112530615 | 0.10 |
ENSG00000242770 |
. |
9168 |
0.23 |
| chr5_88609704_88610064 | 0.10 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
104740 |
0.08 |
| chr5_81682860_81683011 | 0.10 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
81769 |
0.11 |
| chr7_101671263_101671528 | 0.10 |
CTA-357J21.1 |
|
4908 |
0.28 |
| chr9_117440715_117440866 | 0.10 |
ENSG00000272241 |
. |
61963 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.0 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0050922 | negative regulation of chemotaxis(GO:0050922) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.0 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033646 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |