Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
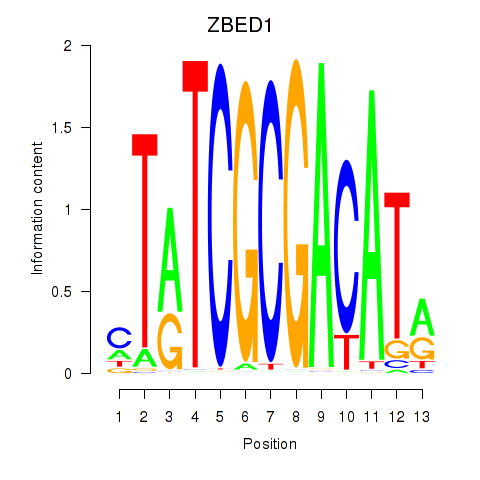
Results for ZBED1
Z-value: 0.29
Transcription factors associated with ZBED1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBED1
|
ENSG00000214717.5 | zinc finger BED-type containing 1 |
|
ZBED1
|
ENSGR0000214717.5 | zinc finger BED-type containing 1 |
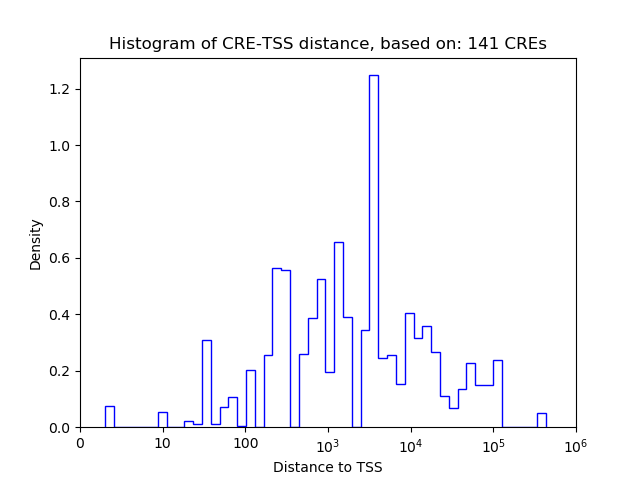
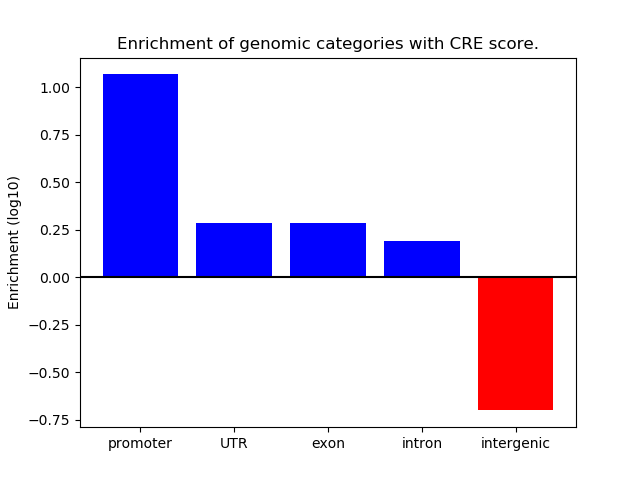
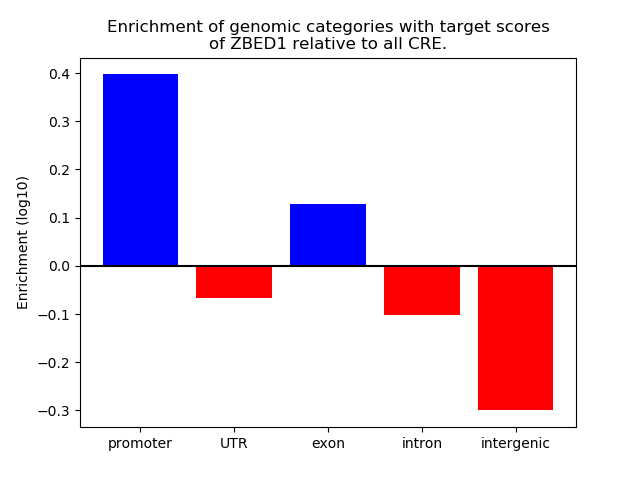
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_2348108_2348259 | ZBED1 | 70413 | 0.105548 | 0.83 | 5.3e-03 | Click! |
| chrX_2348361_2348532 | ZBED1 | 70150 | 0.105982 | 0.47 | 2.0e-01 | Click! |
| chrX_2416147_2416538 | ZBED1 | 2254 | 0.301917 | 0.43 | 2.4e-01 | Click! |
| chrX_2345256_2345407 | ZBED1 | 73265 | 0.100937 | 0.26 | 5.1e-01 | Click! |
| chrX_2416914_2417114 | ZBED1 | 1582 | 0.351667 | 0.15 | 7.0e-01 | Click! |
Activity of the ZBED1 motif across conditions
Conditions sorted by the z-value of the ZBED1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
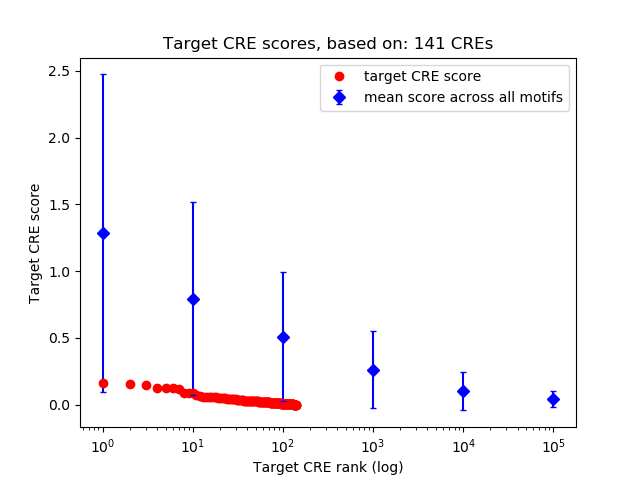
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_25200368_25200588 | 0.17 |
SNURF |
SNRPN upstream reading frame protein |
317 |
0.44 |
| chr22_47017621_47018011 | 0.15 |
GRAMD4 |
GRAM domain containing 4 |
1517 |
0.48 |
| chr17_30822661_30822885 | 0.14 |
RP11-466A19.1 |
|
219 |
0.89 |
| chr4_170541949_170542100 | 0.13 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
36 |
0.98 |
| chr5_140044827_140044997 | 0.13 |
WDR55 |
WD repeat domain 55 |
651 |
0.44 |
| chr14_63999442_63999593 | 0.12 |
PPP2R5E |
protein phosphatase 2, regulatory subunit B', epsilon isoform |
10499 |
0.15 |
| chr8_125736424_125736575 | 0.12 |
MTSS1 |
metastasis suppressor 1 |
3358 |
0.33 |
| chr15_90607365_90607516 | 0.09 |
ZNF710 |
zinc finger protein 710 |
3806 |
0.17 |
| chr10_31321539_31321725 | 0.09 |
ZNF438 |
zinc finger protein 438 |
766 |
0.74 |
| chr14_23235289_23235688 | 0.09 |
OXA1L |
oxidase (cytochrome c) assembly 1-like |
243 |
0.48 |
| chr2_219906286_219906437 | 0.07 |
CCDC108 |
coiled-coil domain containing 108 |
116 |
0.93 |
| chr19_56132593_56132790 | 0.06 |
ZNF784 |
zinc finger protein 784 |
3241 |
0.09 |
| chr3_47516315_47516770 | 0.06 |
SCAP |
SREBF chaperone |
545 |
0.7 |
| chr10_104734953_104735104 | 0.06 |
CNNM2 |
cyclin M2 |
56914 |
0.11 |
| chr1_232665428_232665579 | 0.06 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
14191 |
0.3 |
| chr5_169308444_169308734 | 0.06 |
FAM196B |
family with sequence similarity 196, member B |
99155 |
0.07 |
| chr2_86849098_86849398 | 0.05 |
RNF103-CHMP3 |
RNF103-CHMP3 readthrough |
903 |
0.46 |
| chr1_205181854_205182019 | 0.05 |
DSTYK |
dual serine/threonine and tyrosine protein kinase |
1209 |
0.44 |
| chr17_33387513_33387664 | 0.05 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
3171 |
0.15 |
| chr20_30467211_30467720 | 0.05 |
TTLL9 |
tubulin tyrosine ligase-like family, member 9 |
167 |
0.93 |
| chr3_159572063_159572214 | 0.05 |
SCHIP1 |
schwannomin interacting protein 1 |
1410 |
0.44 |
| chr2_74726093_74726283 | 0.05 |
AC005041.17 |
|
306 |
0.59 |
| chr1_6053839_6054115 | 0.05 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
1206 |
0.38 |
| chr12_48295131_48295282 | 0.04 |
VDR |
vitamin D (1,25- dihydroxyvitamin D3) receptor |
3580 |
0.21 |
| chr14_23292384_23292986 | 0.04 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
65 |
0.91 |
| chr12_865374_865579 | 0.04 |
WNK1 |
WNK lysine deficient protein kinase 1 |
2744 |
0.29 |
| chr1_160694421_160694572 | 0.04 |
CD48 |
CD48 molecule |
12855 |
0.15 |
| chr5_40442466_40442675 | 0.04 |
ENSG00000265615 |
. |
122150 |
0.06 |
| chr8_56802041_56802192 | 0.04 |
RP11-318K15.2 |
|
4038 |
0.17 |
| chr16_56624565_56624716 | 0.04 |
MT3 |
metallothionein 3 |
1188 |
0.28 |
| chr10_21788929_21789081 | 0.04 |
CASC10 |
cancer susceptibility candidate 10 |
2814 |
0.17 |
| chr8_26370627_26370798 | 0.04 |
PNMA2 |
paraneoplastic Ma antigen 2 |
896 |
0.46 |
| chr2_148603094_148603503 | 0.04 |
ACVR2A |
activin A receptor, type IIA |
1113 |
0.55 |
| chr7_80458996_80459147 | 0.04 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
89428 |
0.1 |
| chrX_107335383_107335582 | 0.03 |
ATG4A |
autophagy related 4A, cysteine peptidase |
464 |
0.57 |
| chr14_91861774_91862603 | 0.03 |
CCDC88C |
coiled-coil domain containing 88C |
21502 |
0.21 |
| chr7_66025936_66026087 | 0.03 |
KCTD7 |
potassium channel tetramerization domain containing 7 |
67857 |
0.09 |
| chr3_170625046_170625197 | 0.03 |
EIF5A2 |
eukaryotic translation initiation factor 5A2 |
1266 |
0.51 |
| chr3_57529803_57530341 | 0.03 |
DNAH12 |
dynein, axonemal, heavy chain 12 |
1 |
0.96 |
| chr2_148554372_148554523 | 0.03 |
ACVR2A |
activin A receptor, type IIA |
47639 |
0.15 |
| chr10_22629149_22629300 | 0.03 |
SPAG6 |
sperm associated antigen 6 |
5175 |
0.17 |
| chr12_106532820_106532971 | 0.03 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
916 |
0.65 |
| chr13_36045165_36045339 | 0.03 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
5580 |
0.22 |
| chr5_148929725_148929878 | 0.03 |
CSNK1A1 |
casein kinase 1, alpha 1 |
61 |
0.96 |
| chrX_152927887_152928038 | 0.03 |
ENSG00000244335 |
. |
3719 |
0.13 |
| chr19_36630549_36630790 | 0.03 |
CAPNS1 |
calpain, small subunit 1 |
186 |
0.89 |
| chr12_65600948_65601243 | 0.03 |
LEMD3 |
LEM domain containing 3 |
37744 |
0.16 |
| chr11_32355420_32355641 | 0.03 |
WT1 |
Wilms tumor 1 |
66631 |
0.12 |
| chr14_23630356_23630721 | 0.03 |
ENSG00000202229 |
. |
4915 |
0.12 |
| chr10_14900189_14900340 | 0.03 |
CDNF |
cerebral dopamine neurotrophic factor |
19690 |
0.15 |
| chr21_42738143_42738294 | 0.03 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
3794 |
0.25 |
| chr3_45783013_45783179 | 0.03 |
ENSG00000244357 |
. |
1071 |
0.5 |
| chr5_14129033_14129184 | 0.02 |
TRIO |
trio Rho guanine nucleotide exchange factor |
14703 |
0.3 |
| chr13_113560364_113560515 | 0.02 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
3930 |
0.23 |
| chr11_514795_514946 | 0.02 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
7570 |
0.08 |
| chr10_6579728_6580530 | 0.02 |
PRKCQ |
protein kinase C, theta |
42072 |
0.21 |
| chr5_59284336_59284487 | 0.02 |
CTD-2254N19.1 |
|
32665 |
0.23 |
| chr15_90754517_90755171 | 0.02 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
10076 |
0.12 |
| chr9_127031049_127031200 | 0.02 |
RP11-121A14.3 |
|
5969 |
0.18 |
| chr16_24747560_24747740 | 0.02 |
TNRC6A |
trinucleotide repeat containing 6A |
6616 |
0.28 |
| chr17_16301870_16302159 | 0.02 |
RP11-138I1.4 |
|
16031 |
0.12 |
| chr5_141256597_141256905 | 0.02 |
PCDH1 |
protocadherin 1 |
1203 |
0.48 |
| chr5_76382337_76382809 | 0.02 |
ZBED3-AS1 |
ZBED3 antisense RNA 1 |
8 |
0.88 |
| chr9_81755503_81755654 | 0.02 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
431110 |
0.01 |
| chrX_123124561_123124722 | 0.02 |
STAG2 |
stromal antigen 2 |
27621 |
0.21 |
| chr20_1644368_1644519 | 0.02 |
ENSG00000242348 |
. |
2563 |
0.22 |
| chr3_196471446_196471597 | 0.02 |
ENSG00000272359 |
. |
2674 |
0.18 |
| chr10_35639470_35639863 | 0.02 |
CCNY |
cyclin Y |
13864 |
0.2 |
| chr12_122125138_122125289 | 0.02 |
MORN3 |
MORN repeat containing 3 |
17653 |
0.16 |
| chr4_914129_914321 | 0.02 |
GAK |
cyclin G associated kinase |
4702 |
0.17 |
| chr1_36008630_36009058 | 0.02 |
KIAA0319L |
KIAA0319-like |
11744 |
0.15 |
| chr10_3028303_3028552 | 0.02 |
PFKP |
phosphofructokinase, platelet |
80098 |
0.11 |
| chr1_46908511_46908921 | 0.02 |
RP5-1109J22.1 |
|
10915 |
0.18 |
| chr12_94637273_94637424 | 0.02 |
PLXNC1 |
plexin C1 |
10946 |
0.18 |
| chr16_58617014_58617165 | 0.02 |
CNOT1 |
CCR4-NOT transcription complex, subunit 1 |
8122 |
0.15 |
| chr11_17298613_17298922 | 0.02 |
NUCB2 |
nucleobindin 2 |
197 |
0.95 |
| chr7_152095363_152095514 | 0.01 |
ENSG00000253088 |
. |
4427 |
0.25 |
| chr18_23669463_23669743 | 0.01 |
SS18 |
synovial sarcoma translocation, chromosome 18 |
457 |
0.87 |
| chr3_195150330_195150481 | 0.01 |
ENSG00000207368 |
. |
10130 |
0.19 |
| chr10_112180186_112180405 | 0.01 |
ENSG00000221359 |
. |
13704 |
0.21 |
| chr10_125784070_125784221 | 0.01 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
22096 |
0.24 |
| chr11_1675553_1675820 | 0.01 |
KRTAP5-5 |
keratin associated protein 5-5 |
24653 |
0.09 |
| chr17_76136918_76137344 | 0.01 |
C17orf99 |
chromosome 17 open reading frame 99 |
5303 |
0.12 |
| chr6_74232611_74232913 | 0.01 |
RP11-505P4.7 |
|
579 |
0.47 |
| chr9_127133993_127134300 | 0.01 |
ENSG00000264237 |
. |
21336 |
0.17 |
| chr1_205561220_205561413 | 0.01 |
ENSG00000206762 |
. |
2853 |
0.21 |
| chr18_44526774_44527068 | 0.01 |
KATNAL2 |
katanin p60 subunit A-like 2 |
103 |
0.95 |
| chr14_24025734_24026020 | 0.01 |
THTPA |
thiamine triphosphatase |
331 |
0.49 |
| chr7_65300352_65300503 | 0.01 |
ENSG00000200670 |
. |
20768 |
0.15 |
| chr4_101464634_101464785 | 0.01 |
EMCN |
endomucin |
25459 |
0.26 |
| chr19_40029250_40030224 | 0.01 |
EID2 |
EP300 interacting inhibitor of differentiation 2 |
1133 |
0.32 |
| chr13_60067800_60067951 | 0.01 |
ENSG00000239003 |
. |
13671 |
0.32 |
| chr18_74718576_74718776 | 0.01 |
MBP |
myelin basic protein |
3402 |
0.31 |
| chr10_129786129_129786485 | 0.01 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
699 |
0.77 |
| chr3_10269802_10270219 | 0.01 |
ENSG00000222348 |
. |
11114 |
0.12 |
| chr1_25446820_25447086 | 0.01 |
RP4-706G24.1 |
|
87677 |
0.07 |
| chr8_67940679_67940936 | 0.01 |
PPP1R42 |
protein phosphatase 1, regulatory subunit 42 |
21 |
0.97 |
| chr4_109081453_109081912 | 0.01 |
LEF1 |
lymphoid enhancer-binding factor 1 |
5775 |
0.24 |
| chr10_124608730_124608881 | 0.01 |
CUZD1 |
CUB and zona pellucida-like domains 1 |
1504 |
0.38 |
| chr7_43698266_43698436 | 0.01 |
COA1 |
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
8456 |
0.23 |
| chr4_39700876_39701553 | 0.01 |
UBE2K |
ubiquitin-conjugating enzyme E2K |
1418 |
0.4 |
| chr3_181418656_181418956 | 0.01 |
SOX2-OT |
SOX2 overlapping transcript (non-protein coding) |
1178 |
0.55 |
| chr13_111123516_111123822 | 0.01 |
COL4A2-AS2 |
COL4A2 antisense RNA 2 |
8035 |
0.22 |
| chr4_154713480_154713740 | 0.01 |
SFRP2 |
secreted frizzled-related protein 2 |
3338 |
0.26 |
| chr7_535817_536099 | 0.01 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
22187 |
0.19 |
| chr4_76434081_76434232 | 0.01 |
THAP6 |
THAP domain containing 6 |
5000 |
0.21 |
| chr19_15777024_15777175 | 0.01 |
CYP4F12 |
cytochrome P450, family 4, subfamily F, polypeptide 12 |
6468 |
0.18 |
| chr2_38978217_38978435 | 0.01 |
SRSF7 |
serine/arginine-rich splicing factor 7 |
174 |
0.7 |
| chr12_109197582_109197733 | 0.01 |
SSH1 |
slingshot protein phosphatase 1 |
22405 |
0.13 |
| chr18_46887488_46887730 | 0.01 |
DYM |
dymeclin |
7458 |
0.22 |
| chr4_54927756_54928028 | 0.01 |
AC110792.1 |
HCG2027126; Uncharacterized protein |
679 |
0.65 |
| chr7_102388056_102388207 | 0.01 |
FAM185A |
family with sequence similarity 185, member A |
1344 |
0.44 |
| chr3_85008513_85008834 | 0.00 |
CADM2 |
cell adhesion molecule 2 |
24 |
0.99 |
| chr1_154325026_154325247 | 0.00 |
ENSG00000238365 |
. |
13917 |
0.1 |
| chr20_52789414_52789702 | 0.00 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
630 |
0.79 |
| chr1_36557176_36557327 | 0.00 |
ADPRHL2 |
ADP-ribosylhydrolase like 2 |
2775 |
0.2 |
| chr8_129243121_129243295 | 0.00 |
ENSG00000201782 |
. |
10458 |
0.28 |
| chr12_112546642_112546922 | 0.00 |
NAA25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
44 |
0.97 |
| chr3_72598996_72599234 | 0.00 |
ENSG00000238568 |
. |
23001 |
0.2 |
| chr15_63883849_63884000 | 0.00 |
USP3-AS1 |
USP3 antisense RNA 1 |
2670 |
0.27 |
| chr21_47745359_47745818 | 0.00 |
PCNT |
pericentrin |
1552 |
0.26 |
| chr8_29243512_29244126 | 0.00 |
RP4-676L2.1 |
|
33132 |
0.15 |
| chr17_80263860_80264011 | 0.00 |
CD7 |
CD7 molecule |
11493 |
0.11 |
| chr6_12083621_12083772 | 0.00 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
42283 |
0.2 |
| chr2_33311490_33311641 | 0.00 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
47908 |
0.17 |
| chr12_94206603_94206759 | 0.00 |
ENSG00000264978 |
. |
18171 |
0.19 |
| chr15_40713920_40714255 | 0.00 |
IVD |
isovaleryl-CoA dehydrogenase |
5721 |
0.1 |
| chr18_42788224_42788375 | 0.00 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
4661 |
0.28 |
| chr19_45972792_45973282 | 0.00 |
FOSB |
FBJ murine osteosarcoma viral oncogene homolog B |
97 |
0.94 |
| chr10_28966894_28967045 | 0.00 |
BAMBI |
BMP and activin membrane-bound inhibitor |
698 |
0.68 |
| chr3_100279906_100280202 | 0.00 |
GPR128 |
G protein-coupled receptor 128 |
48379 |
0.14 |
| chr7_86973924_86974573 | 0.00 |
CROT |
carnitine O-octanoyltransferase |
749 |
0.74 |
| chrX_77170418_77170569 | 0.00 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
4294 |
0.18 |
| chr2_101992396_101992547 | 0.00 |
CREG2 |
cellular repressor of E1A-stimulated genes 2 |
11586 |
0.22 |
| chr19_42613495_42613654 | 0.00 |
POU2F2 |
POU class 2 homeobox 2 |
7465 |
0.1 |
| chr20_43272276_43272496 | 0.00 |
ADA |
adenosine deaminase |
7946 |
0.14 |
| chr6_5525547_5525698 | 0.00 |
RP1-232P20.1 |
|
67314 |
0.13 |
| chr5_138641789_138641940 | 0.00 |
MATR3 |
matrin 3 |
1920 |
0.2 |
| chr22_41439766_41439959 | 0.00 |
ENSG00000222698 |
. |
1639 |
0.29 |
| chr22_18172109_18172260 | 0.00 |
BCL2L13 |
BCL2-like 13 (apoptosis facilitator) |
33706 |
0.14 |
| chr3_128878659_128878903 | 0.00 |
ISY1-RAB43 |
ISY1-RAB43 readthrough |
1098 |
0.3 |
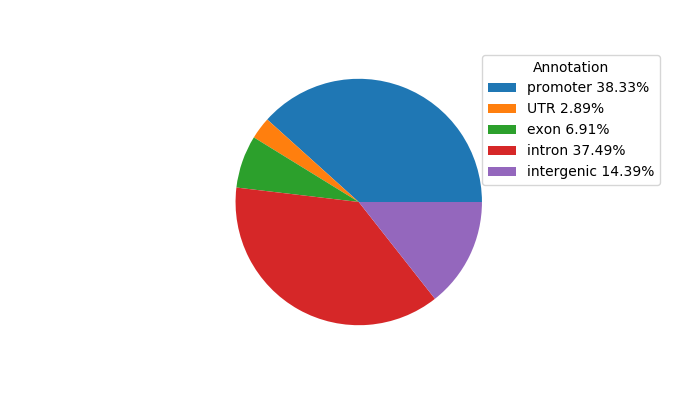
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |