Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
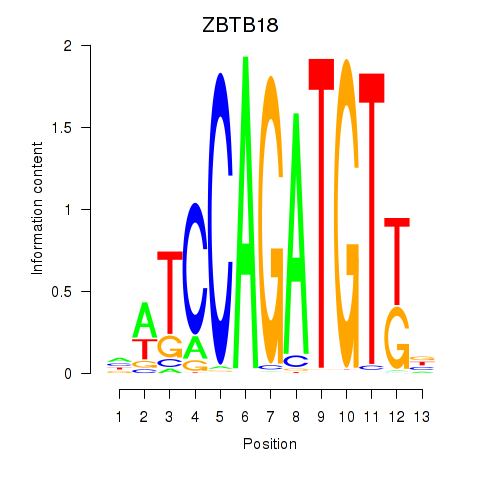
Results for ZBTB18
Z-value: 2.07
Transcription factors associated with ZBTB18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB18
|
ENSG00000179456.9 | zinc finger and BTB domain containing 18 |
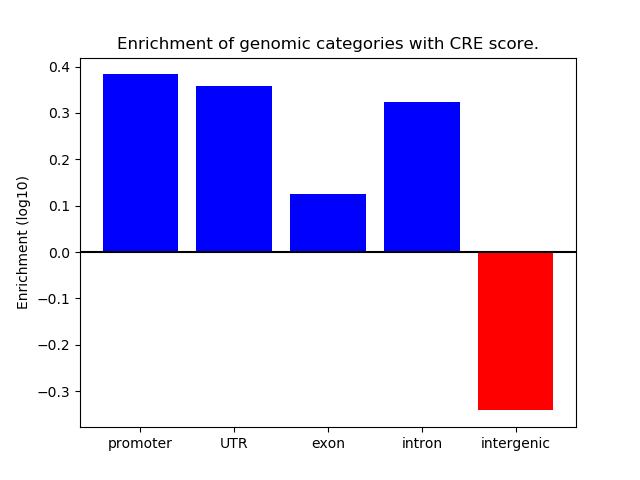
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_244217966_244218117 | ZBTB18 | 3456 | 0.265015 | -0.80 | 9.7e-03 | Click! |
| chr1_244210618_244210958 | ZBTB18 | 3797 | 0.264146 | -0.76 | 1.7e-02 | Click! |
| chr1_244165402_244165576 | ZBTB18 | 49096 | 0.149266 | -0.68 | 4.4e-02 | Click! |
| chr1_244214937_244215088 | ZBTB18 | 427 | 0.859900 | -0.61 | 8.1e-02 | Click! |
| chr1_244215609_244215953 | ZBTB18 | 1196 | 0.521680 | -0.60 | 8.9e-02 | Click! |
Activity of the ZBTB18 motif across conditions
Conditions sorted by the z-value of the ZBTB18 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
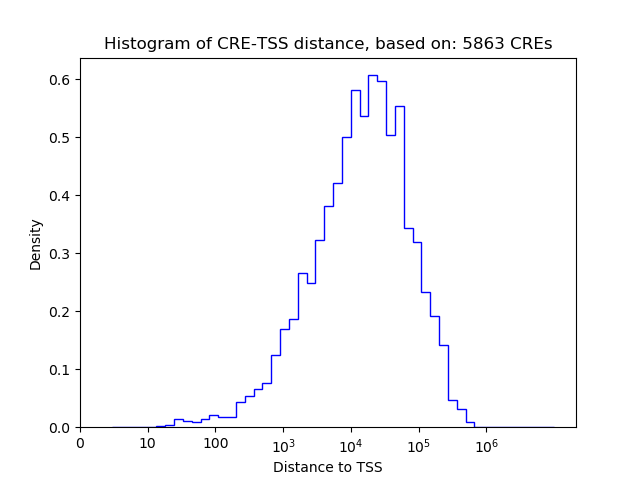
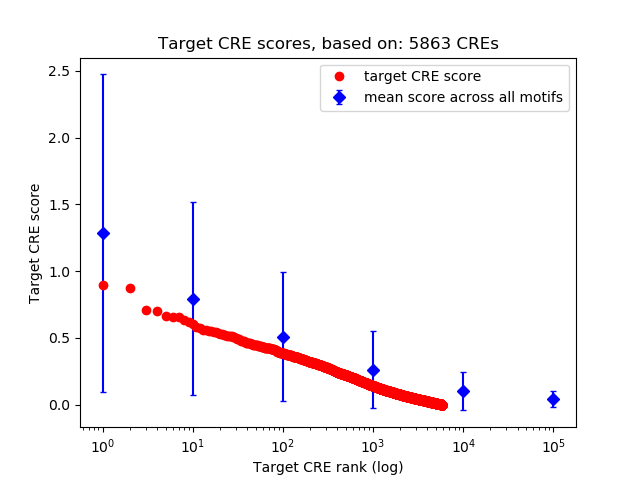
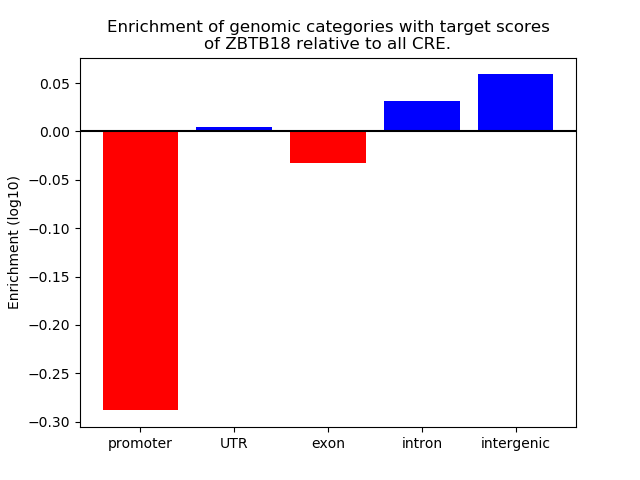
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_40100780_40100931 | 0.90 |
RP1-144F13.3 |
|
1767 |
0.23 |
| chr2_238131211_238131483 | 0.87 |
AC112715.2 |
Uncharacterized protein |
34387 |
0.2 |
| chr17_48343728_48343879 | 0.71 |
TMEM92 |
transmembrane protein 92 |
4964 |
0.14 |
| chr17_71039617_71039918 | 0.70 |
SLC39A11 |
solute carrier family 39, member 11 |
46136 |
0.17 |
| chr12_28721330_28721481 | 0.67 |
CCDC91 |
coiled-coil domain containing 91 |
19408 |
0.26 |
| chr5_56833824_56833975 | 0.66 |
ACTBL2 |
actin, beta-like 2 |
55263 |
0.14 |
| chr19_48674183_48675124 | 0.65 |
C19orf68 |
chromosome 19 open reading frame 68 |
704 |
0.32 |
| chr10_100108984_100109336 | 0.63 |
ENSG00000221419 |
. |
45904 |
0.13 |
| chr11_15178522_15178673 | 0.62 |
INSC |
inscuteable homolog (Drosophila) |
7943 |
0.3 |
| chr20_56622785_56622936 | 0.60 |
ENSG00000221385 |
. |
19652 |
0.22 |
| chr3_98625022_98625173 | 0.59 |
ENSG00000207331 |
. |
2206 |
0.27 |
| chr2_238215371_238215540 | 0.58 |
AC112715.2 |
Uncharacterized protein |
49721 |
0.15 |
| chr11_130608199_130608484 | 0.56 |
C11orf44 |
chromosome 11 open reading frame 44 |
65490 |
0.14 |
| chr21_29827131_29827282 | 0.56 |
ENSG00000251894 |
. |
288324 |
0.01 |
| chr20_3667221_3667372 | 0.55 |
ADAM33 |
ADAM metallopeptidase domain 33 |
4403 |
0.16 |
| chr14_75072847_75072998 | 0.55 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
5853 |
0.22 |
| chr18_10059619_10060204 | 0.54 |
ENSG00000263630 |
. |
54717 |
0.15 |
| chr11_7912101_7912252 | 0.54 |
ENSG00000252769 |
. |
1350 |
0.35 |
| chr8_119499700_119499851 | 0.54 |
SAMD12 |
sterile alpha motif domain containing 12 |
93346 |
0.1 |
| chr6_129507673_129507824 | 0.53 |
ENSG00000252554 |
. |
250377 |
0.02 |
| chr11_74561444_74561595 | 0.53 |
ENSG00000242999 |
. |
4564 |
0.19 |
| chr8_99021155_99021306 | 0.52 |
MATN2 |
matrilin 2 |
7542 |
0.15 |
| chr1_246150406_246150557 | 0.52 |
RP11-83A16.1 |
|
46372 |
0.18 |
| chr2_235934802_235934953 | 0.52 |
SH3BP4 |
SH3-domain binding protein 4 |
31393 |
0.26 |
| chr3_73835223_73835374 | 0.52 |
PDZRN3 |
PDZ domain containing ring finger 3 |
161207 |
0.04 |
| chr2_36657128_36657279 | 0.51 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
73589 |
0.11 |
| chr20_23148002_23148153 | 0.51 |
ENSG00000201527 |
. |
6469 |
0.24 |
| chr14_68943525_68944196 | 0.51 |
RAD51B |
RAD51 paralog B |
65633 |
0.13 |
| chr11_14432940_14433091 | 0.50 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
46963 |
0.15 |
| chr13_114872368_114872623 | 0.50 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
1800 |
0.41 |
| chr11_71698874_71699025 | 0.49 |
IL18BP |
interleukin 18 binding protein |
10638 |
0.11 |
| chr15_49015763_49015914 | 0.49 |
RP11-485O10.2 |
|
59549 |
0.11 |
| chr6_100716336_100716487 | 0.49 |
RP1-121G13.2 |
|
158583 |
0.04 |
| chr17_17780568_17780719 | 0.48 |
TOM1L2 |
target of myb1-like 2 (chicken) |
5265 |
0.19 |
| chr9_101791785_101791940 | 0.47 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
74458 |
0.09 |
| chr11_118492770_118493048 | 0.47 |
AP002954.3 |
|
14429 |
0.11 |
| chr1_44705174_44705804 | 0.47 |
ERI3-IT1 |
ERI3 intronic transcript 1 (non-protein coding) |
4456 |
0.21 |
| chr8_141908612_141908763 | 0.47 |
PTK2 |
protein tyrosine kinase 2 |
7820 |
0.26 |
| chr2_232056745_232056896 | 0.47 |
ARMC9 |
armadillo repeat containing 9 |
6440 |
0.22 |
| chr1_170656810_170656961 | 0.46 |
PRRX1 |
paired related homeobox 1 |
23807 |
0.25 |
| chr2_19740386_19740537 | 0.46 |
OSR1 |
odd-skipped related transciption factor 1 |
182047 |
0.03 |
| chr1_21588856_21589397 | 0.46 |
RP3-329E20.2 |
|
3449 |
0.23 |
| chr2_151332991_151333142 | 0.46 |
RND3 |
Rho family GTPase 3 |
8830 |
0.33 |
| chr8_26519218_26519369 | 0.46 |
DPYSL2 |
dihydropyrimidinase-like 2 |
83372 |
0.1 |
| chr17_12630832_12630983 | 0.45 |
AC005358.1 |
|
4708 |
0.2 |
| chr10_69802673_69802824 | 0.45 |
HERC4 |
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
3024 |
0.27 |
| chr17_1683782_1683933 | 0.45 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
8844 |
0.12 |
| chr9_117508350_117508501 | 0.45 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
45963 |
0.14 |
| chr13_98905870_98906021 | 0.45 |
RP11-573N10.1 |
|
20041 |
0.17 |
| chr15_99881623_99881774 | 0.45 |
AC022819.2 |
Uncharacterized protein |
21870 |
0.18 |
| chr9_82379139_82379290 | 0.45 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
57533 |
0.18 |
| chr12_105812666_105812817 | 0.44 |
C12orf75 |
chromosome 12 open reading frame 75 |
88095 |
0.08 |
| chr10_80916996_80917718 | 0.44 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
88565 |
0.09 |
| chr12_53474857_53475008 | 0.44 |
SPRYD3 |
SPRY domain containing 3 |
1728 |
0.22 |
| chr15_70668335_70668710 | 0.44 |
ENSG00000200216 |
. |
182947 |
0.03 |
| chr7_151297671_151297822 | 0.44 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
31693 |
0.18 |
| chr3_134183959_134184110 | 0.44 |
CEP63 |
centrosomal protein 63kDa |
20551 |
0.16 |
| chr11_44058607_44058758 | 0.43 |
ACCSL |
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional)-like |
10849 |
0.17 |
| chr9_22040392_22040543 | 0.43 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
6283 |
0.2 |
| chr3_88856750_88856901 | 0.43 |
EPHA3 |
EPH receptor A3 |
299849 |
0.01 |
| chr22_27118545_27118696 | 0.43 |
CRYBA4 |
crystallin, beta A4 |
100692 |
0.07 |
| chrY_27544696_27544847 | 0.43 |
ENSG00000272042 |
. |
60283 |
0.14 |
| chr6_144614901_144615052 | 0.43 |
UTRN |
utrophin |
2011 |
0.42 |
| chr19_42785461_42786111 | 0.43 |
CIC |
capicua transcriptional repressor |
2386 |
0.14 |
| chr4_89664108_89664259 | 0.43 |
FAM13A-AS1 |
FAM13A antisense RNA 1 |
21317 |
0.18 |
| chr8_95299657_95299808 | 0.43 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
25154 |
0.22 |
| chrY_26417595_26417746 | 0.43 |
ENSG00000242425 |
. |
60288 |
0.14 |
| chr15_78182901_78183238 | 0.42 |
CSPG4P13 |
chondroitin sulfate proteoglycan 4 pseudogene 13 |
3957 |
0.18 |
| chr12_6658690_6659153 | 0.42 |
IFFO1 |
intermediate filament family orphan 1 |
2145 |
0.13 |
| chr5_65072338_65072489 | 0.42 |
NLN |
neurolysin (metallopeptidase M3 family) |
4830 |
0.27 |
| chr14_52491686_52491966 | 0.42 |
NID2 |
nidogen 2 (osteonidogen) |
13844 |
0.16 |
| chr21_28778719_28778870 | 0.42 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
439962 |
0.01 |
| chr8_95253991_95254142 | 0.42 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
18932 |
0.21 |
| chr7_130012625_130012776 | 0.42 |
CPA1 |
carboxypeptidase A1 (pancreatic) |
7480 |
0.14 |
| chr1_16343988_16344139 | 0.42 |
HSPB7 |
heat shock 27kDa protein family, member 7 (cardiovascular) |
257 |
0.86 |
| chr9_130502702_130502853 | 0.42 |
TOR2A |
torsin family 2, member A |
5173 |
0.1 |
| chr9_89724167_89724318 | 0.42 |
C9orf170 |
chromosome 9 open reading frame 170 |
39317 |
0.21 |
| chr18_56247162_56247463 | 0.42 |
ENSG00000252284 |
. |
20551 |
0.14 |
| chr6_136256944_136257095 | 0.41 |
ENSG00000238921 |
. |
21126 |
0.21 |
| chr9_136271156_136271307 | 0.41 |
REXO4 |
REX4, RNA exonuclease 4 homolog (S. cerevisiae) |
9789 |
0.08 |
| chr17_30662996_30663147 | 0.41 |
C17orf75 |
chromosome 17 open reading frame 75 |
690 |
0.62 |
| chr9_114706444_114706595 | 0.41 |
ENSG00000266315 |
. |
12139 |
0.2 |
| chr17_48718114_48718630 | 0.41 |
ABCC3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
6154 |
0.13 |
| chr14_58312690_58312841 | 0.41 |
ENSG00000252837 |
. |
8387 |
0.24 |
| chr14_88937631_88937782 | 0.41 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
1746 |
0.34 |
| chr14_55057316_55057467 | 0.41 |
SAMD4A |
sterile alpha motif domain containing 4A |
22754 |
0.22 |
| chr1_27730071_27730579 | 0.40 |
GPR3 |
G protein-coupled receptor 3 |
11177 |
0.13 |
| chr1_157992444_157992595 | 0.39 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
2821 |
0.3 |
| chr11_10345196_10345347 | 0.39 |
AMPD3 |
adenosine monophosphate deaminase 3 |
6053 |
0.17 |
| chr8_126582644_126582795 | 0.39 |
ENSG00000266452 |
. |
125912 |
0.06 |
| chr17_66592514_66592665 | 0.39 |
FAM20A |
family with sequence similarity 20, member A |
4941 |
0.26 |
| chr1_214744112_214744263 | 0.39 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
19621 |
0.24 |
| chr15_85888485_85888636 | 0.39 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
35258 |
0.15 |
| chr1_120507427_120507715 | 0.39 |
RP5-1042I8.7 |
|
54335 |
0.13 |
| chr6_140364192_140364343 | 0.39 |
ENSG00000252107 |
. |
115564 |
0.07 |
| chr10_33403329_33403717 | 0.39 |
ENSG00000263576 |
. |
15959 |
0.23 |
| chr3_37154604_37154755 | 0.39 |
ENSG00000206645 |
. |
19954 |
0.15 |
| chr2_73333310_73333461 | 0.39 |
RAB11FIP5 |
RAB11 family interacting protein 5 (class I) |
6761 |
0.18 |
| chr15_74493564_74493715 | 0.39 |
STRA6 |
stimulated by retinoic acid 6 |
1152 |
0.31 |
| chr22_43709368_43709519 | 0.38 |
SCUBE1 |
signal peptide, CUB domain, EGF-like 1 |
29886 |
0.16 |
| chr12_50696750_50697016 | 0.38 |
AC140061.12 |
Uncharacterized protein |
6394 |
0.16 |
| chr19_13127164_13127315 | 0.38 |
CTC-239J10.1 |
|
1914 |
0.18 |
| chr15_48845807_48845958 | 0.38 |
FBN1 |
fibrillin 1 |
92036 |
0.08 |
| chr12_93139497_93139648 | 0.38 |
PLEKHG7 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
9261 |
0.24 |
| chr15_78598378_78598529 | 0.38 |
WDR61 |
WD repeat domain 61 |
6317 |
0.13 |
| chr16_53700893_53701044 | 0.38 |
RPGRIP1L |
RPGRIP1-like |
36754 |
0.17 |
| chr2_12191853_12192027 | 0.38 |
ENSG00000238503 |
. |
21442 |
0.26 |
| chr16_70788132_70788283 | 0.38 |
VAC14-AS1 |
VAC14 antisense RNA 1 |
794 |
0.57 |
| chr7_81254412_81254563 | 0.38 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
137825 |
0.05 |
| chr17_70131143_70131294 | 0.38 |
SOX9 |
SRY (sex determining region Y)-box 9 |
14057 |
0.3 |
| chr5_53843408_53843569 | 0.37 |
SNX18 |
sorting nexin 18 |
29895 |
0.23 |
| chr15_59461408_59461559 | 0.37 |
ENSG00000253030 |
. |
1978 |
0.21 |
| chr4_3709443_3709594 | 0.37 |
ADRA2C |
adrenoceptor alpha 2C |
58557 |
0.14 |
| chr7_55097499_55097650 | 0.37 |
EGFR |
epidermal growth factor receptor |
10763 |
0.3 |
| chr7_134483017_134483168 | 0.37 |
CALD1 |
caldesmon 1 |
18663 |
0.27 |
| chr19_41106420_41106924 | 0.37 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
605 |
0.65 |
| chr11_118475244_118475483 | 0.37 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
1792 |
0.23 |
| chr7_107612741_107612892 | 0.37 |
ENSG00000238297 |
. |
27611 |
0.14 |
| chr1_95484423_95484574 | 0.37 |
RP11-313A24.1 |
|
42654 |
0.12 |
| chr12_96040256_96040407 | 0.37 |
ENSG00000212448 |
. |
28866 |
0.2 |
| chr6_158838381_158838532 | 0.37 |
RP11-732M18.2 |
|
18863 |
0.2 |
| chr6_107039846_107039997 | 0.37 |
ENSG00000200295 |
. |
15670 |
0.17 |
| chr6_143444127_143444389 | 0.37 |
AIG1 |
androgen-induced 1 |
3129 |
0.29 |
| chr22_44453501_44453802 | 0.37 |
PARVB |
parvin, beta |
11290 |
0.25 |
| chr2_232675645_232675796 | 0.37 |
RP11-690I21.2 |
|
1164 |
0.45 |
| chr4_141177038_141177320 | 0.37 |
SCOC |
short coiled-coil protein |
1261 |
0.51 |
| chr12_102821294_102821445 | 0.37 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
51060 |
0.17 |
| chr2_46111015_46111315 | 0.36 |
PRKCE |
protein kinase C, epsilon |
116876 |
0.06 |
| chr16_70672907_70673058 | 0.36 |
IL34 |
interleukin 34 |
7486 |
0.16 |
| chr7_28076131_28076580 | 0.36 |
JAZF1 |
JAZF zinc finger 1 |
15937 |
0.26 |
| chr11_123074983_123075134 | 0.36 |
CLMP |
CXADR-like membrane protein |
9069 |
0.2 |
| chr13_50897665_50897816 | 0.36 |
ENSG00000221198 |
. |
39633 |
0.21 |
| chr15_93571206_93571357 | 0.36 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
19185 |
0.22 |
| chr6_133823044_133823328 | 0.36 |
RP3-323P13.2 |
|
9680 |
0.25 |
| chr8_123101555_123101706 | 0.36 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
447954 |
0.01 |
| chr5_149539504_149539655 | 0.36 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
4156 |
0.17 |
| chr18_42169234_42169525 | 0.36 |
SETBP1 |
SET binding protein 1 |
90759 |
0.1 |
| chr15_50152532_50152683 | 0.36 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
16285 |
0.21 |
| chr11_123060970_123061121 | 0.36 |
CTD-2216M2.1 |
|
1535 |
0.4 |
| chr2_28884288_28884439 | 0.36 |
AC074011.2 |
|
3043 |
0.24 |
| chr1_218549970_218550121 | 0.36 |
TGFB2 |
transforming growth factor, beta 2 |
30468 |
0.19 |
| chr11_19983976_19984127 | 0.36 |
NAV2-AS3 |
NAV2 antisense RNA 3 |
18832 |
0.23 |
| chr1_246024185_246024336 | 0.35 |
RP11-83A16.1 |
|
172593 |
0.03 |
| chr2_189980501_189980652 | 0.35 |
ENSG00000264725 |
. |
17261 |
0.2 |
| chr10_79266628_79266779 | 0.35 |
ENSG00000199592 |
. |
80104 |
0.1 |
| chr22_27908632_27908783 | 0.35 |
RP11-375H17.1 |
|
203761 |
0.03 |
| chr15_41335174_41335443 | 0.35 |
RP11-540O11.4 |
|
18611 |
0.14 |
| chr9_91610255_91610406 | 0.35 |
S1PR3 |
sphingosine-1-phosphate receptor 3 |
1091 |
0.53 |
| chr15_75993395_75993608 | 0.35 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
11688 |
0.09 |
| chr17_79534584_79534839 | 0.35 |
NPLOC4 |
nuclear protein localization 4 homolog (S. cerevisiae) |
1050 |
0.34 |
| chr2_62376728_62376879 | 0.35 |
AC018462.2 |
|
2787 |
0.26 |
| chr1_212655450_212655601 | 0.35 |
NENF |
neudesin neurotrophic factor |
49296 |
0.12 |
| chr9_94533727_94534271 | 0.35 |
ENSG00000266855 |
. |
135466 |
0.05 |
| chr11_28364524_28364785 | 0.35 |
ENSG00000222385 |
. |
81588 |
0.11 |
| chr1_159891458_159892260 | 0.35 |
TAGLN2 |
transgelin 2 |
1648 |
0.22 |
| chr1_241645845_241645996 | 0.35 |
FH |
fumarate hydratase |
37141 |
0.18 |
| chr10_33418933_33419614 | 0.34 |
ENSG00000263576 |
. |
31709 |
0.18 |
| chr6_5989927_5990321 | 0.34 |
NRN1 |
neuritin 1 |
17076 |
0.23 |
| chr6_44041464_44042348 | 0.34 |
RP5-1120P11.1 |
|
483 |
0.79 |
| chr20_17790847_17791092 | 0.34 |
ENSG00000221220 |
. |
32258 |
0.18 |
| chr7_78912243_78912394 | 0.34 |
ENSG00000212482 |
. |
60370 |
0.15 |
| chr6_75853034_75853185 | 0.34 |
COL12A1 |
collagen, type XII, alpha 1 |
8796 |
0.28 |
| chr14_55120501_55120684 | 0.34 |
SAMD4A |
sterile alpha motif domain containing 4A |
85955 |
0.09 |
| chr3_190812380_190812531 | 0.34 |
OSTN |
osteocrin |
104575 |
0.07 |
| chr2_226989431_226989582 | 0.34 |
ENSG00000263363 |
. |
534003 |
0.0 |
| chr1_25132670_25132821 | 0.34 |
ENSG00000238482 |
. |
28275 |
0.19 |
| chr6_46214251_46214402 | 0.34 |
ENPP5 |
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
75618 |
0.1 |
| chr7_91655297_91655448 | 0.34 |
AKAP9 |
A kinase (PRKA) anchor protein 9 |
44080 |
0.16 |
| chr5_140887145_140887296 | 0.34 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
18412 |
0.08 |
| chr12_89842845_89842996 | 0.34 |
POC1B |
POC1 centriolar protein B |
48114 |
0.13 |
| chr9_132379434_132379585 | 0.34 |
C9orf50 |
chromosome 9 open reading frame 50 |
3546 |
0.15 |
| chr5_172225899_172226104 | 0.34 |
ENSG00000206741 |
. |
18904 |
0.13 |
| chr14_31559795_31560006 | 0.34 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
24398 |
0.15 |
| chr2_12957737_12958037 | 0.33 |
ENSG00000264370 |
. |
80394 |
0.11 |
| chr2_196978823_196978974 | 0.33 |
RP11-347P5.1 |
|
37089 |
0.15 |
| chr7_116617261_116617412 | 0.33 |
AC106873.4 |
Uncharacterized protein |
9810 |
0.14 |
| chr6_8084010_8084161 | 0.33 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
13459 |
0.22 |
| chr15_42444184_42444335 | 0.33 |
PLA2G4F |
phospholipase A2, group IVF |
4575 |
0.18 |
| chr7_25887678_25887829 | 0.33 |
ENSG00000199085 |
. |
101853 |
0.08 |
| chr12_92594357_92594508 | 0.33 |
ENSG00000199895 |
. |
30203 |
0.17 |
| chr1_157969918_157970069 | 0.33 |
KIRREL |
kin of IRRE like (Drosophila) |
6558 |
0.23 |
| chr5_168862994_168863145 | 0.33 |
ENSG00000206614 |
. |
116384 |
0.06 |
| chr9_118090029_118090180 | 0.33 |
DEC1 |
deleted in esophageal cancer 1 |
186007 |
0.03 |
| chr10_79804314_79804465 | 0.33 |
RPS24 |
ribosomal protein S24 |
10748 |
0.22 |
| chr6_39521887_39522038 | 0.33 |
KIF6 |
kinesin family member 6 |
42074 |
0.18 |
| chr2_43497360_43497511 | 0.33 |
AC010883.5 |
|
40723 |
0.17 |
| chr1_119544917_119545068 | 0.33 |
TBX15 |
T-box 15 |
12813 |
0.25 |
| chr15_66212983_66213134 | 0.33 |
MEGF11 |
multiple EGF-like-domains 11 |
50500 |
0.12 |
| chr2_182813224_182813375 | 0.32 |
SSFA2 |
sperm specific antigen 2 |
27966 |
0.17 |
| chr22_39828056_39828207 | 0.32 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
25218 |
0.12 |
| chr11_10351462_10351613 | 0.32 |
AMPD3 |
adenosine monophosphate deaminase 3 |
12319 |
0.16 |
| chr16_20963780_20963931 | 0.32 |
LYRM1 |
LYR motif containing 1 |
51429 |
0.11 |
| chr8_97603049_97603490 | 0.32 |
SDC2 |
syndecan 2 |
4654 |
0.31 |
| chr10_101756595_101756746 | 0.32 |
DNMBP |
dynamin binding protein |
13006 |
0.18 |
| chr12_63077564_63077715 | 0.32 |
ENSG00000238475 |
. |
32684 |
0.17 |
| chr19_13957559_13958184 | 0.32 |
ENSG00000207980 |
. |
10398 |
0.08 |
| chr9_15999494_15999645 | 0.32 |
C9orf92 |
chromosome 9 open reading frame 92 |
216328 |
0.02 |
| chr1_18201641_18201961 | 0.32 |
ACTL8 |
actin-like 8 |
119993 |
0.06 |
| chr5_169143698_169143935 | 0.32 |
DOCK2 |
dedicator of cytokinesis 2 |
12863 |
0.26 |
| chr20_4044965_4045249 | 0.32 |
RP11-352D3.2 |
Uncharacterized protein |
10705 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.1 | 0.2 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.2 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.1 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) venous blood vessel development(GO:0060841) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.1 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0010713 | negative regulation of collagen metabolic process(GO:0010713) negative regulation of multicellular organismal metabolic process(GO:0044252) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0035844 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0051957 | positive regulation of amino acid transport(GO:0051957) |
| 0.0 | 0.0 | GO:0009261 | ribonucleotide catabolic process(GO:0009261) |
| 0.0 | 0.5 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.1 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.2 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0032610 | interleukin-1 alpha production(GO:0032610) |
| 0.0 | 0.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.0 | GO:0072160 | nephron tubule epithelial cell differentiation(GO:0072160) regulation of nephron tubule epithelial cell differentiation(GO:0072182) regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.0 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0019883 | antigen processing and presentation of endogenous antigen(GO:0019883) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0032770 | positive regulation of monooxygenase activity(GO:0032770) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.0 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0035268 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0006385 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0071223 | response to lipoteichoic acid(GO:0070391) cellular response to lipoteichoic acid(GO:0071223) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.0 | GO:1904181 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:1903392 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.0 | GO:0060229 | lipase activator activity(GO:0060229) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.7 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.0 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |