Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
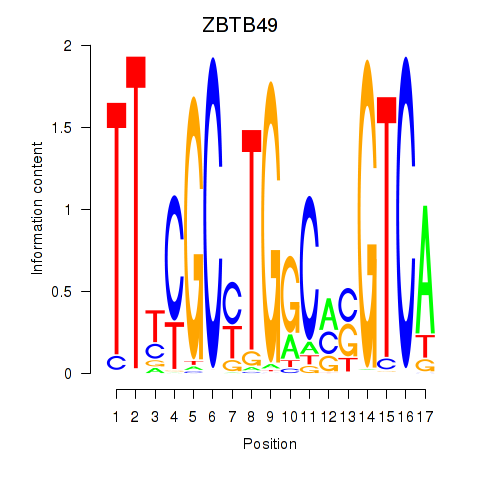
Results for ZBTB49
Z-value: 1.87
Transcription factors associated with ZBTB49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB49
|
ENSG00000168826.11 | zinc finger and BTB domain containing 49 |
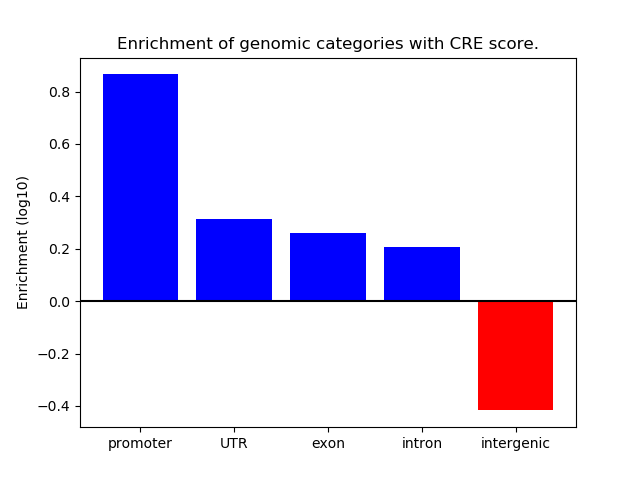
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_4311282_4311433 | ZBTB49 | 7003 | 0.189887 | -0.76 | 1.7e-02 | Click! |
| chr4_4310564_4310715 | ZBTB49 | 6285 | 0.193270 | -0.68 | 4.4e-02 | Click! |
| chr4_4310239_4310390 | ZBTB49 | 5960 | 0.195151 | -0.66 | 5.4e-02 | Click! |
| chr4_4293176_4293389 | ZBTB49 | 1286 | 0.341867 | 0.56 | 1.2e-01 | Click! |
| chr4_4292418_4292569 | ZBTB49 | 497 | 0.515006 | 0.54 | 1.4e-01 | Click! |
Activity of the ZBTB49 motif across conditions
Conditions sorted by the z-value of the ZBTB49 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
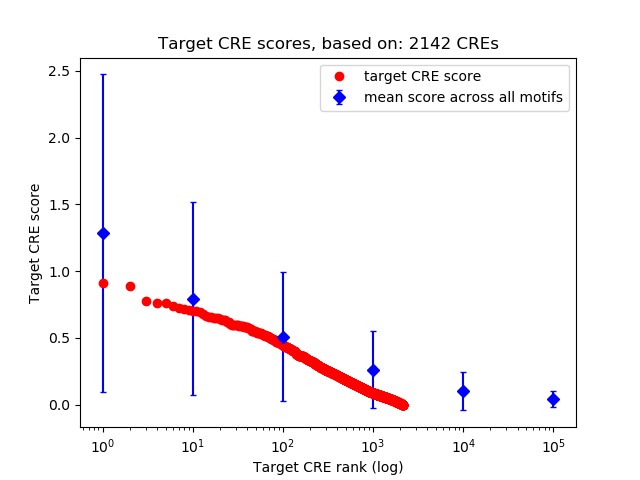
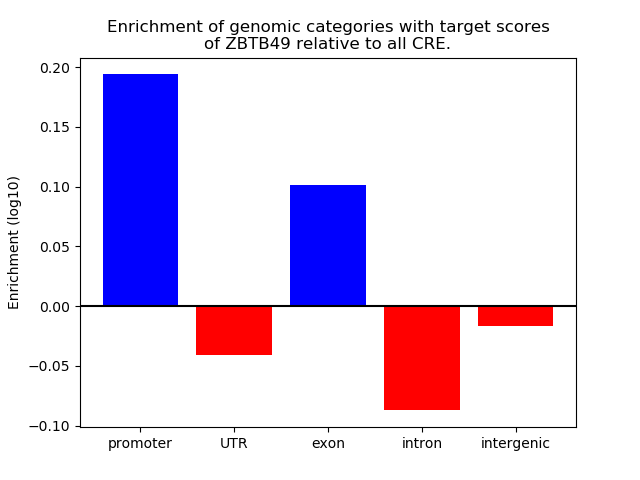
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_35591551_35591702 | 0.91 |
PPP2R3C |
protein phosphatase 2, regulatory subunit B'', gamma |
53 |
0.58 |
| chr5_65440945_65441096 | 0.89 |
SREK1 |
splicing regulatory glutamine/lysine-rich protein 1 |
357 |
0.52 |
| chr1_197879321_197879472 | 0.77 |
LHX9 |
LIM homeobox 9 |
2225 |
0.36 |
| chr11_122714029_122714331 | 0.76 |
CRTAM |
cytotoxic and regulatory T cell molecule |
4972 |
0.24 |
| chr3_194023502_194023858 | 0.76 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
48367 |
0.13 |
| chr1_9764053_9764209 | 0.74 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
6125 |
0.18 |
| chr14_89913899_89914115 | 0.72 |
FOXN3-AS1 |
FOXN3 antisense RNA 1 |
30309 |
0.15 |
| chr1_157819053_157819204 | 0.72 |
CD5L |
CD5 molecule-like |
7540 |
0.21 |
| chr12_47756185_47756391 | 0.71 |
ENSG00000264906 |
. |
1764 |
0.4 |
| chr5_176787986_176788182 | 0.70 |
RGS14 |
regulator of G-protein signaling 14 |
3246 |
0.13 |
| chr9_134377992_134378202 | 0.70 |
POMT1 |
protein-O-mannosyltransferase 1 |
192 |
0.92 |
| chr15_101061308_101061579 | 0.69 |
CERS3 |
ceramide synthase 3 |
23033 |
0.14 |
| chr5_66118075_66118363 | 0.68 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
6383 |
0.33 |
| chr2_220112096_220112414 | 0.66 |
STK16 |
serine/threonine kinase 16 |
1637 |
0.15 |
| chr12_11699871_11700022 | 0.66 |
ENSG00000251747 |
. |
533 |
0.78 |
| chr17_3818792_3818943 | 0.65 |
P2RX1 |
purinergic receptor P2X, ligand-gated ion channel, 1 |
927 |
0.52 |
| chr19_8570015_8570216 | 0.65 |
PRAM1 |
PML-RARA regulated adaptor molecule 1 |
2119 |
0.2 |
| chr6_121758459_121758727 | 0.65 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
1755 |
0.35 |
| chr7_3108313_3108464 | 0.65 |
CARD11 |
caspase recruitment domain family, member 11 |
24809 |
0.21 |
| chr1_26869946_26870282 | 0.64 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
511 |
0.72 |
| chr5_169698686_169698837 | 0.64 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
4430 |
0.26 |
| chr19_14464423_14464642 | 0.63 |
CD97 |
CD97 molecule |
27436 |
0.13 |
| chr3_20084242_20084393 | 0.63 |
KAT2B |
K(lysine) acetyltransferase 2B |
2802 |
0.27 |
| chr11_46371500_46371856 | 0.62 |
DGKZ |
diacylglycerol kinase, zeta |
2540 |
0.22 |
| chr1_182558263_182558414 | 0.62 |
RNASEL |
ribonuclease L (2',5'-oligoisoadenylate synthetase-dependent) |
53 |
0.98 |
| chr1_154988213_154988563 | 0.61 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
1464 |
0.18 |
| chr4_40207735_40207928 | 0.60 |
RHOH |
ras homolog family member H |
5867 |
0.23 |
| chr4_8173290_8173441 | 0.60 |
SH3TC1 |
SH3 domain and tetratricopeptide repeats 1 |
10437 |
0.21 |
| chr22_50752388_50752886 | 0.60 |
XX-C283C717.1 |
|
423 |
0.69 |
| chrX_9542077_9542228 | 0.60 |
TBL1X |
transducin (beta)-like 1X-linked |
39169 |
0.22 |
| chr17_74028313_74028464 | 0.59 |
EVPL |
envoplakin |
4855 |
0.12 |
| chr12_105085856_105086193 | 0.59 |
ENSG00000264295 |
. |
100613 |
0.07 |
| chr3_31274169_31274365 | 0.59 |
ENSG00000222983 |
. |
4060 |
0.33 |
| chr16_50309977_50310263 | 0.59 |
ADCY7 |
adenylate cyclase 7 |
2056 |
0.34 |
| chr11_47237830_47237981 | 0.59 |
DDB2 |
damage-specific DNA binding protein 2, 48kDa |
1412 |
0.25 |
| chr12_65176538_65176711 | 0.59 |
TBC1D30 |
TBC1 domain family, member 30 |
1650 |
0.32 |
| chr7_26373644_26373838 | 0.59 |
SNX10 |
sorting nexin 10 |
19936 |
0.21 |
| chr16_2918250_2918510 | 0.58 |
PRSS22 |
protease, serine, 22 |
10209 |
0.08 |
| chr17_7308499_7308674 | 0.58 |
NLGN2 |
neuroligin 2 |
393 |
0.52 |
| chr20_61604608_61604759 | 0.58 |
SLC17A9 |
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
20285 |
0.14 |
| chr2_233186649_233186800 | 0.57 |
DIS3L2 |
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
11929 |
0.18 |
| chr2_198126272_198126712 | 0.57 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
40751 |
0.13 |
| chr3_183208611_183208762 | 0.57 |
ENSG00000251730 |
. |
36770 |
0.09 |
| chr13_98794878_98795058 | 0.56 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
152 |
0.96 |
| chr14_93672993_93673194 | 0.55 |
C14orf142 |
chromosome 14 open reading frame 142 |
281 |
0.5 |
| chr5_96883161_96883631 | 0.55 |
CTD-2215E18.2 |
|
345812 |
0.01 |
| chr16_88865376_88865527 | 0.55 |
CDT1 |
chromatin licensing and DNA replication factor 1 |
4170 |
0.11 |
| chr11_113847463_113847737 | 0.55 |
HTR3A |
5-hydroxytryptamine (serotonin) receptor 3A, ionotropic |
622 |
0.76 |
| chr22_34317337_34317549 | 0.55 |
LL22NC03-32F9.1 |
|
967 |
0.44 |
| chr22_21323149_21323300 | 0.54 |
AIFM3 |
apoptosis-inducing factor, mitochondrion-associated, 3 |
1134 |
0.29 |
| chr9_102585152_102585422 | 0.54 |
NR4A3 |
nuclear receptor subfamily 4, group A, member 3 |
1150 |
0.6 |
| chr7_40638841_40639130 | 0.54 |
AC004988.1 |
|
52458 |
0.18 |
| chr19_35939401_35939552 | 0.54 |
FFAR2 |
free fatty acid receptor 2 |
273 |
0.86 |
| chr2_242866226_242866494 | 0.54 |
AC131097.3 |
|
9627 |
0.16 |
| chr10_52382609_52382760 | 0.54 |
SGMS1 |
sphingomyelin synthase 1 |
1059 |
0.45 |
| chr4_57775541_57775692 | 0.54 |
REST |
RE1-silencing transcription factor |
1541 |
0.4 |
| chr7_26142036_26142187 | 0.53 |
ENSG00000266430 |
. |
42152 |
0.14 |
| chr9_92035198_92035541 | 0.53 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
1621 |
0.47 |
| chr1_117281810_117282316 | 0.53 |
CD2 |
CD2 molecule |
14944 |
0.19 |
| chr13_97875305_97875640 | 0.52 |
MBNL2 |
muscleblind-like splicing regulator 2 |
863 |
0.74 |
| chr6_159084975_159085126 | 0.52 |
SYTL3 |
synaptotagmin-like 3 |
855 |
0.61 |
| chr8_11142479_11142630 | 0.52 |
MTMR9 |
myotubularin related protein 9 |
49 |
0.97 |
| chr21_46340456_46340607 | 0.52 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
239 |
0.66 |
| chr21_43643624_43643976 | 0.52 |
ENSG00000223262 |
. |
2517 |
0.25 |
| chr1_24117965_24118246 | 0.51 |
LYPLA2 |
lysophospholipase II |
377 |
0.76 |
| chr16_405193_405344 | 0.51 |
AXIN1 |
axin 1 |
2609 |
0.16 |
| chr14_100689570_100689752 | 0.51 |
YY1 |
YY1 transcription factor |
14974 |
0.11 |
| chr22_21195761_21195912 | 0.51 |
PI4KA |
phosphatidylinositol 4-kinase, catalytic, alpha |
17234 |
0.13 |
| chr5_175560441_175560592 | 0.51 |
FAM153B |
family with sequence similarity 153, member B |
48607 |
0.12 |
| chr19_6481975_6482355 | 0.51 |
DENND1C |
DENN/MADD domain containing 1C |
346 |
0.75 |
| chr16_25041687_25041875 | 0.50 |
ARHGAP17 |
Rho GTPase activating protein 17 |
14794 |
0.24 |
| chr4_54231501_54231662 | 0.50 |
SCFD2 |
sec1 family domain containing 2 |
647 |
0.76 |
| chr3_43222048_43222358 | 0.50 |
ENSG00000222331 |
. |
30160 |
0.19 |
| chr18_43923710_43923861 | 0.49 |
RNF165 |
ring finger protein 165 |
9598 |
0.29 |
| chr17_47819458_47819764 | 0.49 |
FAM117A |
family with sequence similarity 117, member A |
17722 |
0.14 |
| chr2_8680555_8680795 | 0.49 |
AC011747.7 |
|
135221 |
0.05 |
| chr1_27940841_27941116 | 0.49 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
9595 |
0.14 |
| chr5_102138051_102138478 | 0.49 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
47217 |
0.2 |
| chr11_128576042_128576248 | 0.49 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
10227 |
0.18 |
| chr9_139000327_139000606 | 0.48 |
C9orf69 |
chromosome 9 open reading frame 69 |
9654 |
0.19 |
| chr4_165878343_165878497 | 0.48 |
FAM218A |
family with sequence similarity 218, member A |
320 |
0.91 |
| chr1_154381143_154381298 | 0.48 |
RP11-350G8.5 |
|
2180 |
0.21 |
| chr7_142012902_142013111 | 0.48 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
23397 |
0.2 |
| chr1_26688110_26688261 | 0.47 |
AIM1L |
absent in melanoma 1-like |
7564 |
0.12 |
| chr12_95186500_95186671 | 0.47 |
ENSG00000208038 |
. |
41589 |
0.19 |
| chr7_37960937_37961095 | 0.47 |
EPDR1 |
ependymin related 1 |
94 |
0.97 |
| chr19_50031302_50031619 | 0.47 |
RCN3 |
reticulocalbin 3, EF-hand calcium binding domain |
87 |
0.9 |
| chr19_51729216_51729410 | 0.47 |
CD33 |
CD33 molecule |
959 |
0.37 |
| chr1_43398456_43398646 | 0.47 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
1755 |
0.36 |
| chr17_29821867_29822018 | 0.47 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
6816 |
0.14 |
| chr7_157112627_157112778 | 0.47 |
ENSG00000266453 |
. |
14215 |
0.22 |
| chr17_75868336_75868487 | 0.47 |
FLJ45079 |
|
10248 |
0.24 |
| chr12_56531942_56532093 | 0.46 |
RP11-603J24.5 |
|
8614 |
0.06 |
| chr1_113589245_113589478 | 0.46 |
LRIG2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
26470 |
0.22 |
| chr1_169664801_169665268 | 0.46 |
SELL |
selectin L |
15805 |
0.19 |
| chr1_206943641_206943792 | 0.46 |
IL10 |
interleukin 10 |
2123 |
0.27 |
| chr16_57578366_57578542 | 0.45 |
GPR114 |
G protein-coupled receptor 114 |
1853 |
0.28 |
| chr8_245965_246116 | 0.45 |
RP5-855D21.2 |
|
58995 |
0.1 |
| chr13_99878346_99878528 | 0.45 |
ENSG00000201793 |
. |
20475 |
0.16 |
| chr9_26956632_26956885 | 0.45 |
IFT74 |
intraflagellar transport 74 homolog (Chlamydomonas) |
281 |
0.65 |
| chr1_206968223_206968416 | 0.44 |
IL19 |
interleukin 19 |
3896 |
0.2 |
| chr16_50699390_50699541 | 0.44 |
RP11-401P9.5 |
|
657 |
0.62 |
| chr1_65627081_65627232 | 0.44 |
AK4 |
adenylate kinase 4 |
13270 |
0.24 |
| chr3_39424652_39424838 | 0.44 |
SLC25A38 |
solute carrier family 25, member 38 |
94 |
0.96 |
| chr17_76254387_76254694 | 0.44 |
TMEM235 |
transmembrane protein 235 |
26418 |
0.12 |
| chr17_62316484_62316635 | 0.43 |
TEX2 |
testis expressed 2 |
8443 |
0.22 |
| chr19_51971617_51971885 | 0.43 |
CEACAM18 |
carcinoembryonic antigen-related cell adhesion molecule 18 |
8087 |
0.1 |
| chr5_76926544_76926695 | 0.43 |
OTP |
orthopedia homeobox |
8894 |
0.23 |
| chr8_145051772_145051990 | 0.43 |
PLEC |
plectin |
979 |
0.36 |
| chrX_39170131_39170563 | 0.43 |
ENSG00000207122 |
. |
229809 |
0.02 |
| chr13_100039643_100039794 | 0.43 |
ENSG00000207719 |
. |
31333 |
0.17 |
| chr17_3818534_3818741 | 0.43 |
P2RX1 |
purinergic receptor P2X, ligand-gated ion channel, 1 |
1157 |
0.44 |
| chr16_67837738_67838160 | 0.43 |
RANBP10 |
RAN binding protein 10 |
2511 |
0.15 |
| chr17_55362857_55363379 | 0.43 |
MSI2 |
musashi RNA-binding protein 2 |
87 |
0.98 |
| chr1_203597474_203597694 | 0.43 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
1656 |
0.46 |
| chr1_154457751_154458027 | 0.42 |
SHE |
Src homology 2 domain containing E |
631 |
0.6 |
| chr16_374247_374531 | 0.42 |
AXIN1 |
axin 1 |
28060 |
0.08 |
| chr3_13236189_13236340 | 0.42 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
121647 |
0.06 |
| chr15_99094347_99094498 | 0.42 |
FAM169B |
family with sequence similarity 169, member B |
36811 |
0.2 |
| chr16_75605342_75605493 | 0.42 |
GABARAPL2 |
GABA(A) receptor-associated protein-like 2 |
4921 |
0.14 |
| chr1_160987971_160988369 | 0.42 |
F11R |
F11 receptor |
2716 |
0.15 |
| chr1_151679471_151679622 | 0.41 |
CELF3 |
CUGBP, Elav-like family member 3 |
840 |
0.36 |
| chr12_4082012_4082345 | 0.41 |
RP11-664D1.1 |
|
67792 |
0.12 |
| chr2_106474919_106475126 | 0.41 |
AC009505.2 |
|
1389 |
0.5 |
| chr1_200983739_200983994 | 0.41 |
KIF21B |
kinesin family member 21B |
8670 |
0.19 |
| chr1_796831_796982 | 0.41 |
AL645608.2 |
|
21137 |
0.12 |
| chr11_111412239_111412489 | 0.41 |
LAYN |
layilin |
87 |
0.95 |
| chr2_234159952_234160103 | 0.41 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
190 |
0.93 |
| chr4_6964522_6964707 | 0.40 |
TBC1D14 |
TBC1 domain family, member 14 |
8667 |
0.14 |
| chr8_24151353_24151528 | 0.40 |
ADAM28 |
ADAM metallopeptidase domain 28 |
113 |
0.98 |
| chr17_3863678_3863829 | 0.40 |
ATP2A3 |
ATPase, Ca++ transporting, ubiquitous |
3832 |
0.22 |
| chr1_155009109_155009453 | 0.40 |
DCST1 |
DC-STAMP domain containing 1 |
2835 |
0.1 |
| chr9_140045822_140045973 | 0.40 |
GRIN1 |
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
11964 |
0.06 |
| chr11_48102714_48102946 | 0.40 |
ENSG00000263693 |
. |
15504 |
0.22 |
| chr2_101949101_101949252 | 0.40 |
ENSG00000264857 |
. |
23264 |
0.16 |
| chr11_117822226_117822384 | 0.40 |
TMPRSS13 |
transmembrane protease, serine 13 |
22131 |
0.15 |
| chr4_109351683_109351834 | 0.39 |
ENSG00000266046 |
. |
146538 |
0.04 |
| chr13_34318080_34318631 | 0.39 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
73831 |
0.12 |
| chr9_117690184_117690335 | 0.39 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
2438 |
0.4 |
| chr11_58347260_58347411 | 0.38 |
ZFP91 |
ZFP91 zinc finger protein |
751 |
0.54 |
| chr5_95194711_95194982 | 0.38 |
ENSG00000250362 |
. |
2644 |
0.2 |
| chr3_71703355_71703506 | 0.38 |
FOXP1 |
forkhead box P1 |
70290 |
0.09 |
| chr11_74662937_74663088 | 0.38 |
SPCS2 |
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
2443 |
0.19 |
| chr18_47824525_47824817 | 0.38 |
CXXC1 |
CXXC finger protein 1 |
9997 |
0.15 |
| chr11_64126806_64126957 | 0.37 |
RPS6KA4 |
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
125 |
0.92 |
| chr9_104211779_104211948 | 0.37 |
ALDOB |
aldolase B, fructose-bisphosphate |
13758 |
0.14 |
| chr7_129908667_129908818 | 0.37 |
CPA2 |
carboxypeptidase A2 (pancreatic) |
2075 |
0.25 |
| chr17_73566405_73566556 | 0.37 |
MYO15B |
myosin XVB pseudogene |
20378 |
0.1 |
| chr9_131124320_131124475 | 0.37 |
URM1 |
ubiquitin related modifier 1 |
9201 |
0.1 |
| chrX_78420820_78421003 | 0.37 |
GPR174 |
G protein-coupled receptor 174 |
5558 |
0.35 |
| chr8_58907122_58907322 | 0.37 |
FAM110B |
family with sequence similarity 110, member B |
109 |
0.98 |
| chr2_219993360_219993640 | 0.37 |
ENSG00000240317 |
. |
26344 |
0.08 |
| chr1_182759043_182759194 | 0.37 |
NPL |
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
207 |
0.95 |
| chr17_46102923_46103182 | 0.37 |
COPZ2 |
coatomer protein complex, subunit zeta 2 |
8186 |
0.09 |
| chr6_11380234_11380472 | 0.37 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
2179 |
0.43 |
| chr17_79005380_79005531 | 0.37 |
BAIAP2 |
BAI1-associated protein 2 |
3495 |
0.16 |
| chr19_55136229_55136380 | 0.37 |
ENSG00000264799 |
. |
4610 |
0.12 |
| chr11_4111133_4111394 | 0.37 |
RRM1 |
ribonucleotide reductase M1 |
4776 |
0.22 |
| chr1_229797401_229797552 | 0.37 |
URB2 |
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
10882 |
0.21 |
| chr10_99894914_99895070 | 0.37 |
R3HCC1L |
R3H domain and coiled-coil containing 1-like |
564 |
0.83 |
| chr9_95792049_95792266 | 0.37 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
14815 |
0.18 |
| chr19_17862547_17862728 | 0.36 |
FCHO1 |
FCH domain only 1 |
292 |
0.86 |
| chr5_54523316_54523467 | 0.36 |
MCIDAS |
multiciliate differentiation and DNA synthesis associated cell cycle protein |
248 |
0.88 |
| chr2_102914112_102914534 | 0.36 |
IL1RL1 |
interleukin 1 receptor-like 1 |
13639 |
0.19 |
| chr4_53525877_53526028 | 0.36 |
USP46 |
ubiquitin specific peptidase 46 |
450 |
0.84 |
| chr7_37734454_37734605 | 0.36 |
GPR141 |
G protein-coupled receptor 141 |
11054 |
0.23 |
| chr5_112054668_112054870 | 0.36 |
APC |
adenomatous polyposis coli |
11550 |
0.24 |
| chr8_99954932_99955099 | 0.36 |
STK3 |
serine/threonine kinase 3 |
40 |
0.97 |
| chr5_143549883_143550034 | 0.36 |
YIPF5 |
Yip1 domain family, member 5 |
236 |
0.7 |
| chr16_69166899_69167129 | 0.36 |
CIRH1A |
cirrhosis, autosomal recessive 1A (cirhin) |
227 |
0.74 |
| chr9_139515902_139516065 | 0.36 |
ENSG00000252440 |
. |
19016 |
0.09 |
| chr20_1111581_1111955 | 0.35 |
PSMF1 |
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
4053 |
0.25 |
| chr8_10588360_10588511 | 0.35 |
SOX7 |
SRY (sex determining region Y)-box 7 |
413 |
0.74 |
| chr5_55626039_55626389 | 0.35 |
ENSG00000210678 |
. |
32698 |
0.18 |
| chr21_39844669_39844820 | 0.35 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
25601 |
0.26 |
| chr1_19991027_19991230 | 0.35 |
HTR6 |
5-hydroxytryptamine (serotonin) receptor 6, G protein-coupled |
652 |
0.69 |
| chr2_8450496_8450864 | 0.35 |
AC011747.7 |
|
365216 |
0.01 |
| chr12_105074262_105074425 | 0.35 |
ENSG00000264295 |
. |
88932 |
0.08 |
| chr12_93322034_93322185 | 0.35 |
EEA1 |
early endosome antigen 1 |
998 |
0.61 |
| chr1_28451935_28452285 | 0.35 |
ENSG00000253005 |
. |
17242 |
0.12 |
| chr5_159826406_159826584 | 0.34 |
C5orf54 |
chromosome 5 open reading frame 54 |
548 |
0.71 |
| chrX_9926593_9926744 | 0.34 |
AC002365.1 |
Homo sapiens uncharacterized LOC100288814 (LOC100288814), mRNA. |
8724 |
0.21 |
| chr3_50312444_50312595 | 0.34 |
LSMEM2 |
leucine-rich single-pass membrane protein 2 |
3939 |
0.09 |
| chr14_98637898_98638078 | 0.34 |
ENSG00000222066 |
. |
160099 |
0.04 |
| chr6_16634011_16634287 | 0.34 |
RP1-151F17.1 |
|
127220 |
0.06 |
| chr10_114501840_114502050 | 0.34 |
RP11-25C19.3 |
|
64367 |
0.12 |
| chr5_55904489_55904655 | 0.34 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
2513 |
0.36 |
| chr1_154968975_154969126 | 0.34 |
LENEP |
lens epithelial protein |
2986 |
0.1 |
| chr2_198169813_198170140 | 0.34 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
2733 |
0.23 |
| chr17_72588151_72588338 | 0.34 |
CD300LD |
CD300 molecule-like family member d |
178 |
0.91 |
| chr2_71692941_71693092 | 0.34 |
DYSF |
dysferlin |
816 |
0.72 |
| chr3_196669893_196670052 | 0.33 |
NCBP2 |
nuclear cap binding protein subunit 2, 20kDa |
504 |
0.69 |
| chr2_149309885_149310036 | 0.33 |
MBD5 |
methyl-CpG binding domain protein 5 |
83666 |
0.1 |
| chr2_2690206_2690393 | 0.33 |
ENSG00000263570 |
. |
330803 |
0.01 |
| chr3_30653249_30653701 | 0.33 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
5382 |
0.33 |
| chr9_97402176_97402371 | 0.33 |
FBP1 |
fructose-1,6-bisphosphatase 1 |
146 |
0.97 |
| chr6_53155154_53155631 | 0.33 |
ENSG00000264056 |
. |
13601 |
0.2 |
| chr12_105043947_105044098 | 0.33 |
ENSG00000264295 |
. |
58611 |
0.13 |
| chr5_131778755_131778906 | 0.33 |
AC116366.5 |
|
16424 |
0.12 |
| chr6_91182223_91182574 | 0.33 |
ENSG00000252676 |
. |
25907 |
0.24 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 0.3 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.3 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.2 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.1 | 0.3 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.1 | 0.2 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.1 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.3 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.1 | 0.4 | GO:0045217 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.2 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.1 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.4 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.2 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.1 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.2 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.1 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0002524 | hypersensitivity(GO:0002524) regulation of hypersensitivity(GO:0002883) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0001820 | serotonin secretion(GO:0001820) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.2 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0006168 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.8 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0002857 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.7 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.4 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.4 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.0 | GO:0032661 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0051882 | mitochondrial depolarization(GO:0051882) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.2 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.0 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.1 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.3 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.3 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.2 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.2 | GO:0042772 | DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.2 | GO:0045061 | thymic T cell selection(GO:0045061) |
| 0.0 | 0.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:2001257 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) regulation of calcium ion transmembrane transporter activity(GO:1901019) regulation of cation channel activity(GO:2001257) |
| 0.0 | 0.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.0 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.0 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0006560 | proline metabolic process(GO:0006560) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.0 | GO:0043552 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.0 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.3 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.0 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.0 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.0 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.0 | GO:0010954 | positive regulation of protein processing(GO:0010954) positive regulation of protein maturation(GO:1903319) |
| 0.0 | 0.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.0 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.0 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.0 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.2 | GO:1990266 | neutrophil chemotaxis(GO:0030593) neutrophil migration(GO:1990266) |
| 0.0 | 0.0 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0071445 | obsolete cellular response to protein stimulus(GO:0071445) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.7 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.0 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.2 | GO:0019674 | NAD metabolic process(GO:0019674) |
| 0.0 | 0.0 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.0 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.1 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.5 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.0 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.3 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.3 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.6 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.7 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.4 | GO:0016502 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.0 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.0 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.0 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.2 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |