Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
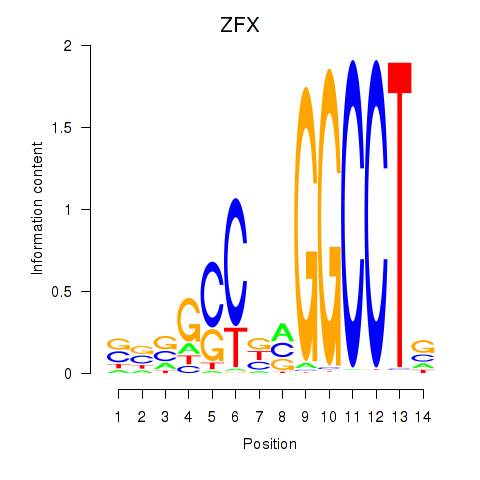
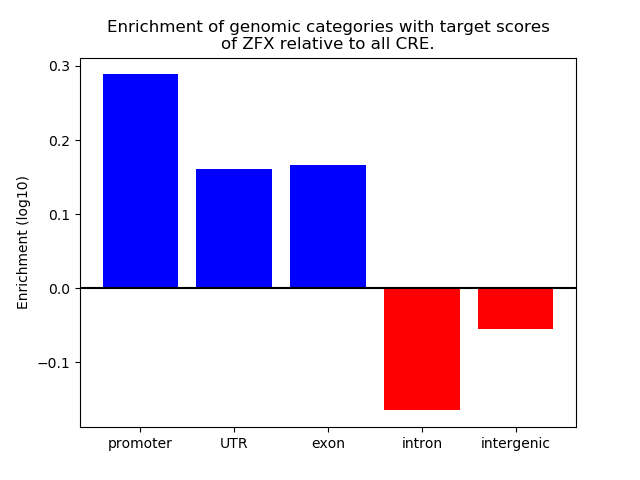
Results for ZFX
Z-value: 2.36
Transcription factors associated with ZFX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFX
|
ENSG00000005889.11 | zinc finger protein X-linked |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_24169836_24170019 | ZFX | 48 | 0.969668 | -0.73 | 2.6e-02 | Click! |
| chrX_24225476_24225627 | ZFX | 34719 | 0.158774 | 0.65 | 6.1e-02 | Click! |
| chrX_24169541_24169817 | ZFX | 200 | 0.924335 | -0.60 | 8.6e-02 | Click! |
| chrX_24166137_24167088 | ZFX | 1134 | 0.354384 | -0.59 | 9.7e-02 | Click! |
| chrX_24169146_24169493 | ZFX | 560 | 0.685146 | -0.58 | 1.0e-01 | Click! |
Activity of the ZFX motif across conditions
Conditions sorted by the z-value of the ZFX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
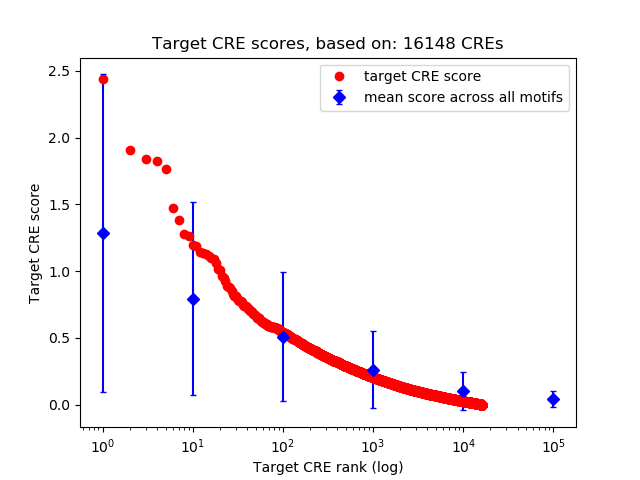
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_76740387_76741175 | 2.44 |
SALL3 |
spalt-like transcription factor 3 |
506 |
0.87 |
| chr10_133999367_134000256 | 1.91 |
RP11-140A10.3 |
|
345 |
0.67 |
| chr1_895413_895840 | 1.84 |
KLHL17 |
kelch-like family member 17 |
341 |
0.68 |
| chr12_4140465_4141124 | 1.82 |
RP11-664D1.1 |
|
126408 |
0.05 |
| chr18_55094933_55095260 | 1.76 |
ONECUT2 |
one cut homeobox 2 |
7821 |
0.21 |
| chr1_22259868_22260816 | 1.47 |
HSPG2 |
heparan sulfate proteoglycan 2 |
3448 |
0.18 |
| chr13_114831463_114832357 | 1.38 |
RASA3 |
RAS p21 protein activator 3 |
11528 |
0.24 |
| chr7_157405725_157406364 | 1.28 |
AC005481.5 |
Uncharacterized protein |
671 |
0.75 |
| chr9_140093664_140094251 | 1.26 |
TPRN |
taperin |
600 |
0.41 |
| chr15_57612815_57613005 | 1.20 |
TCF12 |
transcription factor 12 |
38257 |
0.16 |
| chr3_47844781_47845806 | 1.19 |
DHX30 |
DEAH (Asp-Glu-Ala-His) box helicase 30 |
638 |
0.69 |
| chr17_79486044_79486605 | 1.15 |
RP13-766D20.2 |
|
258 |
0.83 |
| chr19_13318401_13318895 | 1.14 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
49566 |
0.08 |
| chr16_203573_203998 | 1.13 |
HBZ |
hemoglobin, zeta |
1099 |
0.28 |
| chr6_137365603_137366227 | 1.11 |
IL20RA |
interleukin 20 receptor, alpha |
186 |
0.96 |
| chr19_1168901_1169806 | 1.10 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
411 |
0.76 |
| chr16_214202_214642 | 1.09 |
HBM |
hemoglobin, mu |
1555 |
0.19 |
| chr14_24837858_24838637 | 1.06 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
41 |
0.94 |
| chr1_9473459_9474229 | 1.01 |
ENSG00000252956 |
. |
23993 |
0.2 |
| chr19_1409918_1411040 | 1.01 |
DAZAP1 |
DAZ associated protein 1 |
2745 |
0.1 |
| chr22_37914494_37915327 | 0.97 |
CARD10 |
caspase recruitment domain family, member 10 |
337 |
0.83 |
| chr1_228268932_228269720 | 0.95 |
ARF1 |
ADP-ribosylation factor 1 |
1035 |
0.41 |
| chr9_140171055_140171601 | 0.93 |
TOR4A |
torsin family 4, member A |
873 |
0.34 |
| chr10_22725352_22725818 | 0.89 |
RP11-301N24.3 |
|
75484 |
0.09 |
| chr2_241856671_241856922 | 0.88 |
AC104809.3 |
Protein LOC728763 |
1406 |
0.36 |
| chr17_40913377_40913932 | 0.87 |
RAMP2 |
receptor (G protein-coupled) activity modifying protein 2 |
378 |
0.71 |
| chr14_103242729_103243285 | 0.85 |
TRAF3 |
TNF receptor-associated factor 3 |
806 |
0.63 |
| chr20_3183761_3184370 | 0.83 |
DDRGK1 |
DDRGK domain containing 1 |
1266 |
0.33 |
| chr9_140117151_140117759 | 0.82 |
RNF208 |
ring finger protein 208 |
1422 |
0.14 |
| chr11_118780857_118781283 | 0.81 |
ENSG00000264211 |
. |
347 |
0.56 |
| chr8_21766211_21767399 | 0.80 |
DOK2 |
docking protein 2, 56kDa |
4369 |
0.22 |
| chr1_44412862_44413792 | 0.78 |
IPO13 |
importin 13 |
716 |
0.44 |
| chr11_118481078_118481549 | 0.78 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
2955 |
0.16 |
| chr19_47524229_47524951 | 0.78 |
NPAS1 |
neuronal PAS domain protein 1 |
447 |
0.8 |
| chr4_1003833_1004600 | 0.77 |
FGFRL1 |
fibroblast growth factor receptor-like 1 |
449 |
0.75 |
| chrX_200229_200380 | 0.75 |
PLCXD1 |
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
97 |
0.98 |
| chr5_398631_398914 | 0.74 |
AHRR |
aryl-hydrocarbon receptor repressor |
22263 |
0.14 |
| chr19_15559479_15560330 | 0.74 |
ENSG00000269782 |
. |
455 |
0.47 |
| chr11_61311867_61312018 | 0.74 |
LRRC10B |
leucine rich repeat containing 10B |
35670 |
0.11 |
| chr2_8675294_8675837 | 0.73 |
AC011747.7 |
|
140331 |
0.05 |
| chr1_246952409_246952662 | 0.72 |
ENSG00000227953 |
. |
1391 |
0.38 |
| chr10_129535281_129535534 | 0.72 |
FOXI2 |
forkhead box I2 |
92 |
0.98 |
| chr14_60982288_60982505 | 0.70 |
C14orf39 |
chromosome 14 open reading frame 39 |
135 |
0.86 |
| chr20_62694983_62695936 | 0.70 |
TCEA2 |
transcription elongation factor A (SII), 2 |
19 |
0.95 |
| chr1_1293641_1294187 | 0.70 |
MXRA8 |
matrix-remodelling associated 8 |
1 |
0.93 |
| chr6_35181746_35182466 | 0.69 |
SCUBE3 |
signal peptide, CUB domain, EGF-like 3 |
84 |
0.97 |
| chr17_40673744_40674238 | 0.68 |
ENSG00000265611 |
. |
7785 |
0.08 |
| chr8_21646968_21647258 | 0.68 |
GFRA2 |
GDNF family receptor alpha 2 |
348 |
0.92 |
| chr9_139971203_139971717 | 0.67 |
UAP1L1 |
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 |
493 |
0.49 |
| chr8_145014050_145014477 | 0.66 |
PLEC |
plectin |
505 |
0.66 |
| chr5_60921647_60922495 | 0.65 |
C5orf64 |
chromosome 5 open reading frame 64 |
11565 |
0.22 |
| chr17_79455442_79455852 | 0.65 |
ENSG00000266077 |
. |
22488 |
0.08 |
| chr11_34196090_34196535 | 0.65 |
ENSG00000201867 |
. |
14409 |
0.25 |
| chr6_393086_393625 | 0.65 |
IRF4 |
interferon regulatory factor 4 |
1616 |
0.51 |
| chr17_38501915_38502668 | 0.65 |
RARA |
retinoic acid receptor, alpha |
798 |
0.45 |
| chr20_34203648_34204152 | 0.63 |
SPAG4 |
sperm associated antigen 4 |
86 |
0.95 |
| chr14_101308246_101309004 | 0.63 |
ENSG00000214548 |
. |
7020 |
0.04 |
| chr8_143807685_143808400 | 0.63 |
CTD-2292P10.4 |
|
349 |
0.54 |
| chr5_149895200_149895351 | 0.62 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
7601 |
0.18 |
| chr17_80010173_80011309 | 0.62 |
GPS1 |
G protein pathway suppressor 1 |
632 |
0.4 |
| chr17_1686320_1687263 | 0.61 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
11778 |
0.11 |
| chr17_73074006_73074769 | 0.61 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
9435 |
0.08 |
| chr19_3950919_3951410 | 0.61 |
DAPK3 |
death-associated protein kinase 3 |
9993 |
0.1 |
| chr6_39789659_39789810 | 0.61 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
26608 |
0.21 |
| chr3_45589006_45589723 | 0.60 |
LARS2-AS1 |
LARS2 antisense RNA 1 |
38327 |
0.14 |
| chr9_140352686_140353315 | 0.60 |
NSMF |
NMDA receptor synaptonuclear signaling and neuronal migration factor |
754 |
0.52 |
| chr11_61276806_61277546 | 0.60 |
LRRC10B |
leucine rich repeat containing 10B |
904 |
0.38 |
| chr9_139518377_139518866 | 0.59 |
ENSG00000252440 |
. |
21654 |
0.09 |
| chr2_43201843_43202268 | 0.59 |
ENSG00000207087 |
. |
116577 |
0.06 |
| chr19_30334052_30334510 | 0.59 |
CCNE1 |
cyclin E1 |
26171 |
0.23 |
| chr17_43221225_43221687 | 0.59 |
HEXIM1 |
hexamethylene bis-acetamide inducible 1 |
3228 |
0.13 |
| chr9_140317981_140318404 | 0.59 |
NOXA1 |
NADPH oxidase activator 1 |
345 |
0.55 |
| chr12_662501_663191 | 0.59 |
B4GALNT3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
10547 |
0.16 |
| chr1_63783066_63783708 | 0.59 |
RP4-792G4.2 |
|
3813 |
0.21 |
| chr20_49348254_49348561 | 0.58 |
PARD6B |
par-6 family cell polarity regulator beta |
205 |
0.94 |
| chr16_75278250_75278858 | 0.58 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
1954 |
0.25 |
| chr16_88838432_88838925 | 0.58 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
12941 |
0.08 |
| chr6_35393359_35394053 | 0.58 |
FANCE |
Fanconi anemia, complementation group E |
26432 |
0.15 |
| chr19_46380030_46380314 | 0.58 |
IRF2BP1 |
interferon regulatory factor 2 binding protein 1 |
9204 |
0.1 |
| chr14_61104414_61104775 | 0.58 |
SIX1 |
SIX homeobox 1 |
11586 |
0.21 |
| chr10_80827843_80828373 | 0.58 |
ZMIZ1-AS1 |
ZMIZ1 antisense RNA 1 |
456 |
0.62 |
| chr3_57198236_57199040 | 0.58 |
IL17RD |
interleukin 17 receptor D |
765 |
0.66 |
| chr11_3159025_3159707 | 0.57 |
OSBPL5 |
oxysterol binding protein-like 5 |
8807 |
0.17 |
| chr16_49315668_49315968 | 0.57 |
CBLN1 |
cerebellin 1 precursor |
76 |
0.98 |
| chr14_65224545_65225258 | 0.57 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
20148 |
0.18 |
| chr1_6239594_6239889 | 0.57 |
CHD5 |
chromodomain helicase DNA binding protein 5 |
442 |
0.76 |
| chr11_326229_326864 | 0.57 |
IFITM3 |
interferon induced transmembrane protein 3 |
991 |
0.28 |
| chr6_10426428_10427175 | 0.57 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
6930 |
0.18 |
| chr7_25891719_25892347 | 0.57 |
ENSG00000199085 |
. |
97573 |
0.09 |
| chr2_25078291_25078442 | 0.57 |
ADCY3 |
adenylate cyclase 3 |
13861 |
0.17 |
| chr5_132166329_132166761 | 0.56 |
SHROOM1 |
shroom family member 1 |
45 |
0.96 |
| chr9_138952025_138952443 | 0.56 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
9808 |
0.22 |
| chr2_118981232_118982329 | 0.56 |
INSIG2 |
insulin induced gene 2 |
135730 |
0.05 |
| chr11_65321440_65321876 | 0.55 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
460 |
0.63 |
| chr17_75475212_75475601 | 0.55 |
SEPT9 |
septin 9 |
2586 |
0.23 |
| chr11_65486527_65487369 | 0.55 |
KAT5 |
K(lysine) acetyltransferase 5 |
771 |
0.41 |
| chr12_52540643_52541115 | 0.55 |
ENSG00000265804 |
. |
33170 |
0.09 |
| chr22_38614803_38615828 | 0.55 |
MAFF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
5773 |
0.12 |
| chr6_37616181_37616595 | 0.55 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
2238 |
0.4 |
| chr11_8102991_8103508 | 0.54 |
TUB |
tubby bipartite transcription factor |
340 |
0.86 |
| chr1_41130861_41131209 | 0.54 |
RIMS3 |
regulating synaptic membrane exocytosis 3 |
82 |
0.97 |
| chr17_76875045_76875748 | 0.54 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
5164 |
0.16 |
| chr17_62708693_62709583 | 0.54 |
RP13-104F24.1 |
|
45583 |
0.12 |
| chr1_3652951_3653102 | 0.54 |
TP73-AS1 |
TP73 antisense RNA 1 |
1413 |
0.28 |
| chr2_241391852_241392233 | 0.54 |
GPC1 |
glypican 1 |
186 |
0.93 |
| chr15_31691736_31691887 | 0.53 |
KLF13 |
Kruppel-like factor 13 |
33454 |
0.23 |
| chr2_27295479_27295811 | 0.53 |
OST4 |
oligosaccharyltransferase 4 homolog (S. cerevisiae) |
1004 |
0.27 |
| chr7_98029982_98030440 | 0.53 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
169 |
0.97 |
| chr10_120788305_120788993 | 0.53 |
NANOS1 |
nanos homolog 1 (Drosophila) |
579 |
0.67 |
| chr9_137366473_137366866 | 0.53 |
RXRA |
retinoid X receptor, alpha |
68241 |
0.11 |
| chr17_80551323_80552124 | 0.53 |
RP13-638C3.3 |
|
5436 |
0.11 |
| chr12_48207204_48207904 | 0.53 |
HDAC7 |
histone deacetylase 7 |
287 |
0.89 |
| chr20_32272946_32273379 | 0.53 |
E2F1 |
E2F transcription factor 1 |
1048 |
0.35 |
| chr19_46285643_46285794 | 0.52 |
DMPK |
dystrophia myotonica-protein kinase |
18 |
0.94 |
| chr22_46439284_46440015 | 0.52 |
RP6-109B7.5 |
|
9324 |
0.11 |
| chr19_42785461_42786111 | 0.52 |
CIC |
capicua transcriptional repressor |
2386 |
0.14 |
| chr11_1147635_1147786 | 0.51 |
MUC5AC |
mucin 5AC, oligomeric mucus/gel-forming |
3870 |
0.23 |
| chr19_33716510_33717125 | 0.51 |
SLC7A10 |
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10 |
61 |
0.96 |
| chr9_18906751_18906902 | 0.51 |
ENSG00000223211 |
. |
71901 |
0.09 |
| chr16_29466841_29467271 | 0.51 |
SULT1A4 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
71 |
0.8 |
| chr2_149894989_149895325 | 0.51 |
LYPD6B |
LY6/PLAUR domain containing 6B |
75 |
0.99 |
| chr8_145543248_145543399 | 0.51 |
GS1-393G12.12 |
|
4522 |
0.09 |
| chr22_46403312_46403647 | 0.50 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
30470 |
0.1 |
| chr1_156675699_156676565 | 0.50 |
CRABP2 |
cellular retinoic acid binding protein 2 |
524 |
0.6 |
| chr2_241899813_241900212 | 0.50 |
AC104809.3 |
Protein LOC728763 |
1214 |
0.3 |
| chr7_6895095_6895550 | 0.50 |
ENSG00000263457 |
. |
22941 |
0.16 |
| chr7_495260_495411 | 0.50 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
62810 |
0.12 |
| chr19_35630229_35630677 | 0.49 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
108 |
0.92 |
| chr1_17287544_17287908 | 0.49 |
MFAP2 |
microfibrillar-associated protein 2 |
17045 |
0.13 |
| chr1_225934809_225934960 | 0.49 |
ENSG00000223306 |
. |
5907 |
0.16 |
| chr8_49341467_49342503 | 0.49 |
ENSG00000252710 |
. |
121395 |
0.06 |
| chr5_170736823_170737111 | 0.49 |
TLX3 |
T-cell leukemia homeobox 3 |
679 |
0.69 |
| chr4_2463884_2464333 | 0.49 |
RNF4 |
ring finger protein 4 |
161 |
0.92 |
| chr17_17585251_17585813 | 0.49 |
RAI1 |
retinoic acid induced 1 |
277 |
0.91 |
| chr5_178003954_178004239 | 0.49 |
COL23A1 |
collagen, type XXIII, alpha 1 |
13460 |
0.2 |
| chr19_14584769_14585060 | 0.49 |
PTGER1 |
prostaglandin E receptor 1 (subtype EP1), 42kDa |
1260 |
0.29 |
| chr15_40339457_40340106 | 0.49 |
SRP14 |
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
8392 |
0.16 |
| chr11_46274872_46275023 | 0.49 |
CTD-2589M5.4 |
|
21151 |
0.17 |
| chr7_27260020_27260307 | 0.49 |
HOTTIP |
HOXA distal transcript antisense RNA |
20107 |
0.07 |
| chr11_73069874_73070025 | 0.49 |
ENSG00000266866 |
. |
1769 |
0.28 |
| chr13_100620685_100621184 | 0.48 |
ZIC5 |
Zic family member 5 |
3229 |
0.24 |
| chr16_70463887_70464888 | 0.48 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
8604 |
0.12 |
| chr15_40633164_40633428 | 0.48 |
C15orf52 |
chromosome 15 open reading frame 52 |
128 |
0.9 |
| chr8_145690043_145690705 | 0.48 |
CYHR1 |
cysteine/histidine-rich 1 |
44 |
0.88 |
| chr16_19179620_19179966 | 0.48 |
SYT17 |
synaptotagmin XVII |
211 |
0.94 |
| chr17_7819497_7820171 | 0.48 |
AC025335.1 |
|
563 |
0.56 |
| chr8_140716343_140716851 | 0.47 |
KCNK9 |
potassium channel, subfamily K, member 9 |
1298 |
0.63 |
| chr7_100859763_100860679 | 0.47 |
ZNHIT1 |
zinc finger, HIT-type containing 1 |
728 |
0.34 |
| chr6_1821074_1821225 | 0.47 |
FOXC1 |
forkhead box C1 |
210468 |
0.02 |
| chr1_223936103_223936684 | 0.47 |
CAPN2 |
calpain 2, (m/II) large subunit |
36359 |
0.16 |
| chr1_875869_876619 | 0.47 |
SAMD11 |
sterile alpha motif domain containing 11 |
1589 |
0.22 |
| chr2_65085563_65085984 | 0.47 |
ENSG00000199964 |
. |
24482 |
0.15 |
| chr5_415533_415708 | 0.46 |
AHRR |
aryl-hydrocarbon receptor repressor |
5415 |
0.17 |
| chr9_110399429_110399916 | 0.46 |
ENSG00000202308 |
. |
25855 |
0.23 |
| chr13_30084221_30084372 | 0.46 |
MTUS2-AS1 |
MTUS2 antisense RNA 1 |
20054 |
0.24 |
| chr19_11590864_11591191 | 0.46 |
ELAVL3 |
ELAV like neuron-specific RNA binding protein 3 |
413 |
0.69 |
| chrX_152990547_152991011 | 0.46 |
ABCD1 |
ATP-binding cassette, sub-family D (ALD), member 1 |
456 |
0.5 |
| chr5_148785713_148785864 | 0.46 |
ENSG00000208035 |
. |
22693 |
0.11 |
| chr2_74875100_74875395 | 0.46 |
M1AP |
meiosis 1 associated protein |
83 |
0.96 |
| chr19_46877189_46877483 | 0.46 |
AC007193.9 |
|
9090 |
0.12 |
| chr10_112837601_112838021 | 0.46 |
ADRA2A |
adrenoceptor alpha 2A |
1021 |
0.64 |
| chr15_99489885_99490412 | 0.46 |
IGF1R |
insulin-like growth factor 1 receptor |
22319 |
0.19 |
| chr2_236579045_236579338 | 0.46 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
943 |
0.69 |
| chr9_132223080_132223572 | 0.46 |
ENSG00000264298 |
. |
17509 |
0.21 |
| chr3_129335401_129335596 | 0.45 |
PLXND1 |
plexin D1 |
9837 |
0.17 |
| chr16_89258494_89259013 | 0.45 |
SLC22A31 |
solute carrier family 22, member 31 |
9319 |
0.12 |
| chr1_6301763_6302398 | 0.45 |
HES3 |
hes family bHLH transcription factor 3 |
2172 |
0.18 |
| chr14_90990119_90990270 | 0.45 |
ENSG00000252748 |
. |
41291 |
0.16 |
| chrX_47433764_47434046 | 0.45 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
7807 |
0.13 |
| chr10_101604205_101604725 | 0.45 |
DNMBP |
dynamin binding protein |
53554 |
0.11 |
| chr11_116705965_116706733 | 0.45 |
APOA1-AS |
APOA1 antisense RNA |
484 |
0.63 |
| chr12_52585063_52585443 | 0.45 |
KRT80 |
keratin 80 |
531 |
0.69 |
| chr15_41785353_41785895 | 0.44 |
ITPKA |
inositol-trisphosphate 3-kinase A |
33 |
0.97 |
| chr14_61747742_61748391 | 0.44 |
TMEM30B |
transmembrane protein 30B |
107 |
0.63 |
| chr19_932497_932724 | 0.44 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
101 |
0.92 |
| chr3_52124413_52124839 | 0.44 |
LINC00696 |
long intergenic non-protein coding RNA 696 |
27059 |
0.11 |
| chr19_12666271_12666422 | 0.44 |
ZNF564 |
zinc finger protein 564 |
3990 |
0.13 |
| chr10_4868611_4868968 | 0.44 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
301 |
0.93 |
| chr9_116106360_116106630 | 0.44 |
WDR31 |
WD repeat domain 31 |
3917 |
0.15 |
| chr15_68627387_68627538 | 0.44 |
FEM1B |
fem-1 homolog b (C. elegans) |
44912 |
0.15 |
| chr10_131749932_131750083 | 0.44 |
EBF3 |
early B-cell factor 3 |
12098 |
0.3 |
| chr13_114898126_114898522 | 0.44 |
RASA3 |
RAS p21 protein activator 3 |
238 |
0.94 |
| chr1_46998384_46998918 | 0.43 |
MKNK1-AS1 |
MKNK1 antisense RNA 1 |
5717 |
0.16 |
| chr3_47050686_47051097 | 0.43 |
NBEAL2 |
neurobeachin-like 2 |
4120 |
0.22 |
| chr16_88518586_88519370 | 0.43 |
ZFPM1 |
zinc finger protein, FOG family member 1 |
747 |
0.63 |
| chr2_20818076_20818344 | 0.43 |
HS1BP3 |
HCLS1 binding protein 3 |
6323 |
0.22 |
| chr10_29168723_29168874 | 0.43 |
ENSG00000199402 |
. |
5362 |
0.27 |
| chr11_1145070_1145742 | 0.43 |
MUC5AC |
mucin 5AC, oligomeric mucus/gel-forming |
6174 |
0.2 |
| chr10_105379098_105379249 | 0.43 |
RP11-416N2.4 |
|
10797 |
0.17 |
| chr1_36788360_36788868 | 0.43 |
RP11-268J15.5 |
|
721 |
0.48 |
| chr11_702778_703267 | 0.43 |
EPS8L2 |
EPS8-like 2 |
2195 |
0.14 |
| chr7_4848752_4849021 | 0.43 |
ENSG00000206895 |
. |
2034 |
0.27 |
| chr11_33721547_33721982 | 0.43 |
C11orf91 |
chromosome 11 open reading frame 91 |
179 |
0.94 |
| chr4_8474079_8474524 | 0.43 |
RP11-689P11.3 |
|
17632 |
0.17 |
| chr18_44773846_44774145 | 0.42 |
SKOR2 |
SKI family transcriptional corepressor 2 |
1559 |
0.4 |
| chrX_152992299_152993172 | 0.42 |
ABCD1 |
ATP-binding cassette, sub-family D (ALD), member 1 |
1463 |
0.23 |
| chr19_14192503_14193024 | 0.42 |
C19orf67 |
chromosome 19 open reading frame 67 |
3811 |
0.11 |
| chr19_45755493_45756253 | 0.42 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
1323 |
0.34 |
| chr5_1445460_1445611 | 0.42 |
SLC6A3 |
solute carrier family 6 (neurotransmitter transporter), member 3 |
10 |
0.98 |
| chr22_22291490_22292290 | 0.42 |
LL22NC03-86G7.1 |
|
719 |
0.44 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.2 | 0.8 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 1.6 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 0.8 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.2 | 0.6 | GO:0033622 | integrin activation(GO:0033622) |
| 0.2 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 0.6 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.2 | 0.5 | GO:0032328 | alanine transport(GO:0032328) |
| 0.2 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.4 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 0.4 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 0.4 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.1 | 0.3 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.4 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.7 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.1 | 0.7 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.1 | 0.2 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.1 | 0.3 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.1 | 0.4 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 0.1 | GO:0061515 | erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.1 | 0.2 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.5 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 0.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.1 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.3 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.1 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.2 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.2 | GO:0060433 | bronchus development(GO:0060433) |
| 0.1 | 0.2 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.1 | 0.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.1 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0071883 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.3 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.1 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.1 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.2 | GO:0050686 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.1 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.1 | 0.2 | GO:0046084 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.2 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.1 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 0.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.1 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.5 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.1 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.1 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.1 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.8 | GO:1901222 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 0.2 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.2 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.1 | 0.2 | GO:0017000 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.1 | 0.1 | GO:0042637 | catagen(GO:0042637) |
| 0.1 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.1 | GO:0032329 | serine transport(GO:0032329) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.8 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 1.0 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) neural crest cell fate commitment(GO:0014034) |
| 0.0 | 0.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.5 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.2 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.0 | GO:0032616 | interleukin-13 production(GO:0032616) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:0072191 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.4 | GO:0048194 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.0 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.0 | GO:0060896 | neural plate pattern specification(GO:0060896) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.2 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.2 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.0 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.2 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.3 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.2 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.0 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.2 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.2 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0032069 | regulation of nuclease activity(GO:0032069) |
| 0.0 | 0.0 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.1 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.3 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 0.0 | GO:1903579 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.3 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 1.3 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0051458 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.3 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.1 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.3 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.1 | GO:0043030 | regulation of macrophage activation(GO:0043030) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.2 | GO:0048895 | sensory system development(GO:0048880) lateral line nerve development(GO:0048892) lateral line nerve glial cell differentiation(GO:0048895) lateral line system development(GO:0048925) lateral line nerve glial cell development(GO:0048937) iridophore differentiation(GO:0050935) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.0 | GO:0097300 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.0 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.1 | GO:0090504 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.0 | 0.3 | GO:0032648 | regulation of interferon-beta production(GO:0032648) |
| 0.0 | 0.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.3 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.0 | 0.1 | GO:0043032 | positive regulation of macrophage activation(GO:0043032) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0021772 | olfactory bulb development(GO:0021772) |
| 0.0 | 0.1 | GO:2000193 | regulation of icosanoid secretion(GO:0032303) positive regulation of icosanoid secretion(GO:0032305) regulation of prostaglandin secretion(GO:0032306) positive regulation of prostaglandin secretion(GO:0032308) prostaglandin secretion(GO:0032310) positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0043555 | regulation of translation in response to stress(GO:0043555) regulation of translational initiation in response to stress(GO:0043558) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0061365 | positive regulation of lipoprotein lipase activity(GO:0051006) positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0051956 | negative regulation of amino acid transport(GO:0051956) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0071397 | cellular response to sterol(GO:0036315) cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.0 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.1 | GO:0040018 | positive regulation of multicellular organism growth(GO:0040018) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0051608 | histamine secretion(GO:0001821) histamine transport(GO:0051608) |
| 0.0 | 0.0 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0090559 | regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.1 | GO:0042517 | positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 | 0.0 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 0.0 | GO:0043576 | regulation of respiratory gaseous exchange(GO:0043576) |
| 0.0 | 0.0 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0042559 | folic acid-containing compound biosynthetic process(GO:0009396) pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.2 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.0 | 0.0 | GO:1903514 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.0 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.0 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.0 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.0 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.0 | 0.1 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.2 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.1 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.1 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.0 | 0.1 | GO:0051828 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.0 | GO:0032376 | positive regulation of sterol transport(GO:0032373) positive regulation of cholesterol transport(GO:0032376) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.0 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.0 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.0 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0001974 | blood vessel remodeling(GO:0001974) |
| 0.0 | 0.2 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.0 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:2000573 | positive regulation of DNA biosynthetic process(GO:2000573) |
| 0.0 | 0.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0045684 | positive regulation of epidermal cell differentiation(GO:0045606) positive regulation of epidermis development(GO:0045684) |
| 0.0 | 0.0 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.0 | GO:0032608 | interferon-beta production(GO:0032608) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.5 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0030277 | epithelial structure maintenance(GO:0010669) maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0032449 | CBM complex(GO:0032449) |
| 0.2 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 1.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.8 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.5 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.0 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.6 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.0 | 0.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.2 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 4.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.0 | 0.5 | GO:0044309 | neuron spine(GO:0044309) |
| 0.0 | 0.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 0.5 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.4 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.5 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 0.7 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.3 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 1.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.5 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 0.7 | GO:0046970 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.2 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.1 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.5 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 2.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.3 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.2 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.0 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) purine nucleobase binding(GO:0002060) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.0 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.0 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.0 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.1 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.6 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |