Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
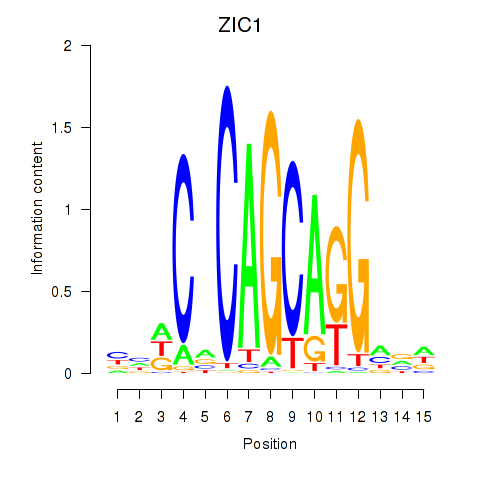
Results for ZIC1
Z-value: 1.58
Transcription factors associated with ZIC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC1
|
ENSG00000152977.5 | Zic family member 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_147130089_147130240 | ZIC1 | 2993 | 0.245780 | -0.68 | 4.3e-02 | Click! |
| chr3_147136818_147136969 | ZIC1 | 9722 | 0.191752 | 0.64 | 6.3e-02 | Click! |
| chr3_147130587_147130738 | ZIC1 | 3491 | 0.230377 | -0.55 | 1.2e-01 | Click! |
| chr3_147110427_147110578 | ZIC1 | 1180 | 0.453421 | 0.52 | 1.5e-01 | Click! |
| chr3_147111345_147111496 | ZIC1 | 262 | 0.909593 | 0.51 | 1.7e-01 | Click! |
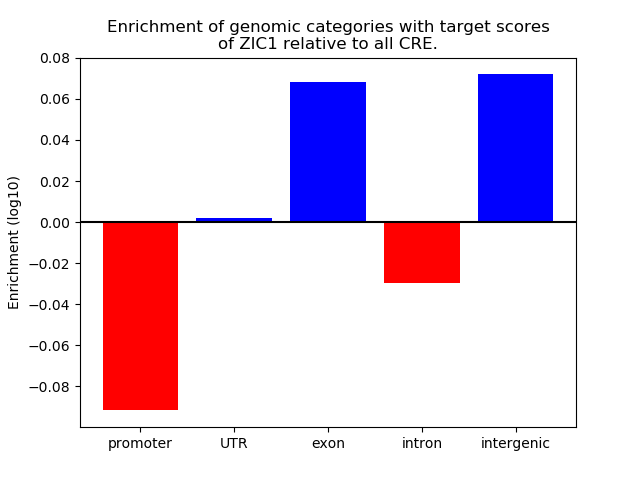
Activity of the ZIC1 motif across conditions
Conditions sorted by the z-value of the ZIC1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
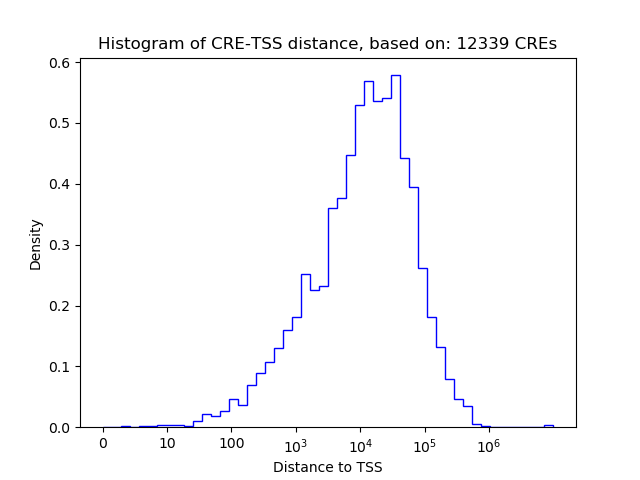
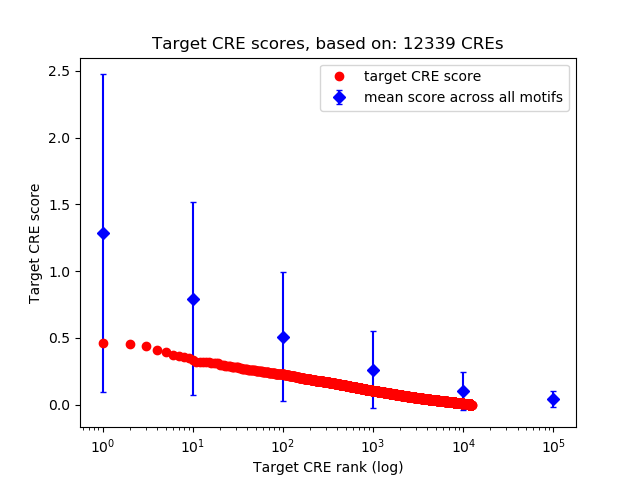
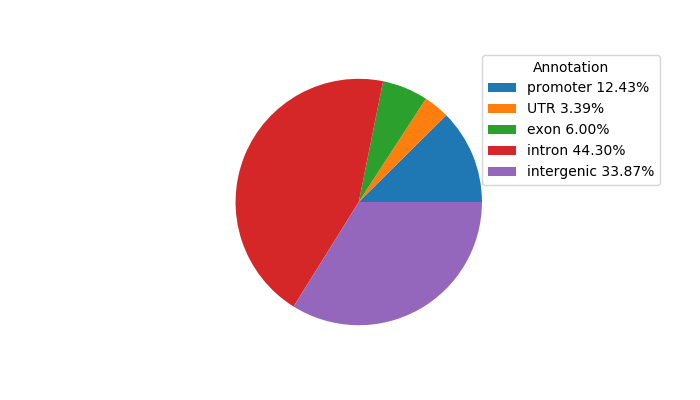
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_21441508_21442003 | 0.46 |
LAMA3 |
laminin, alpha 3 |
11049 |
0.22 |
| chr5_1494557_1494933 | 0.45 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
29347 |
0.18 |
| chr1_109934550_109935130 | 0.44 |
SORT1 |
sortilin 1 |
1139 |
0.45 |
| chr12_43709758_43710210 | 0.41 |
ENSG00000215993 |
. |
24601 |
0.27 |
| chr14_75735835_75736333 | 0.39 |
RP11-293M10.1 |
Uncharacterized protein |
98 |
0.95 |
| chr4_124399950_124400344 | 0.37 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
79024 |
0.12 |
| chr7_48009158_48009388 | 0.37 |
HUS1 |
HUS1 checkpoint homolog (S. pombe) |
9828 |
0.19 |
| chr16_53127962_53128318 | 0.36 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
4932 |
0.27 |
| chr5_73536989_73537227 | 0.35 |
ENSG00000222551 |
. |
30482 |
0.23 |
| chr15_70440007_70440428 | 0.34 |
ENSG00000200216 |
. |
45358 |
0.16 |
| chr4_37666460_37666702 | 0.32 |
RELL1 |
RELT-like 1 |
422 |
0.87 |
| chr1_150136184_150136335 | 0.32 |
PLEKHO1 |
pleckstrin homology domain containing, family O member 1 |
7136 |
0.15 |
| chr20_23132592_23133523 | 0.32 |
ENSG00000201527 |
. |
8551 |
0.22 |
| chr19_41926852_41927085 | 0.32 |
BCKDHA |
branched chain keto acid dehydrogenase E1, alpha polypeptide |
2046 |
0.16 |
| chr14_61568522_61568874 | 0.32 |
SLC38A6 |
solute carrier family 38, member 6 |
58830 |
0.13 |
| chr11_65259042_65259542 | 0.32 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
33256 |
0.08 |
| chr7_117468416_117468567 | 0.31 |
CTTNBP2 |
cortactin binding protein 2 |
36657 |
0.22 |
| chr12_66289441_66289878 | 0.31 |
RP11-366L20.2 |
Uncharacterized protein |
13712 |
0.17 |
| chr9_71173939_71174218 | 0.31 |
TMEM252 |
transmembrane protein 252 |
18295 |
0.25 |
| chr11_12129395_12129640 | 0.30 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
2621 |
0.38 |
| chr6_16773666_16773859 | 0.30 |
RP1-151F17.1 |
|
11619 |
0.24 |
| chr12_81102661_81102850 | 0.30 |
MYF6 |
myogenic factor 6 (herculin) |
1478 |
0.44 |
| chr7_41046808_41047238 | 0.29 |
AC005160.3 |
|
231866 |
0.02 |
| chr11_115801802_115801953 | 0.29 |
ENSG00000239153 |
. |
303974 |
0.01 |
| chr19_935300_935451 | 0.29 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
2866 |
0.12 |
| chr18_19904044_19904557 | 0.29 |
ENSG00000238907 |
. |
63761 |
0.11 |
| chr19_50284218_50285014 | 0.29 |
AP2A1 |
adaptor-related protein complex 2, alpha 1 subunit |
14225 |
0.07 |
| chr2_172380569_172381265 | 0.29 |
CYBRD1 |
cytochrome b reductase 1 |
1328 |
0.53 |
| chr16_67168849_67169000 | 0.28 |
C16orf70 |
chromosome 16 open reading frame 70 |
11263 |
0.08 |
| chr8_38874641_38875005 | 0.28 |
ENSG00000207199 |
. |
1379 |
0.37 |
| chr8_141129055_141129276 | 0.28 |
C8orf17 |
chromosome 8 open reading frame 17 |
185749 |
0.03 |
| chr10_110225723_110225990 | 0.28 |
ENSG00000222436 |
. |
475212 |
0.01 |
| chr1_212106101_212106282 | 0.28 |
ENSG00000212187 |
. |
92712 |
0.06 |
| chr18_61498762_61499118 | 0.27 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
39986 |
0.14 |
| chr15_68992418_68992757 | 0.27 |
CORO2B |
coronin, actin binding protein, 2B |
68260 |
0.12 |
| chr1_45296298_45296558 | 0.27 |
PTCH2 |
patched 2 |
7496 |
0.09 |
| chr2_20795415_20795848 | 0.27 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
3323 |
0.28 |
| chr1_229161136_229161564 | 0.26 |
RP5-1061H20.5 |
|
201959 |
0.02 |
| chr11_10620459_10620776 | 0.26 |
MRVI1-AS1 |
MRVI1 antisense RNA 1 |
5873 |
0.19 |
| chr3_52904617_52904846 | 0.26 |
TMEM110 |
transmembrane protein 110 |
18016 |
0.12 |
| chr1_245215838_245216124 | 0.26 |
ENSG00000251754 |
. |
8751 |
0.17 |
| chr6_6728512_6728665 | 0.26 |
LY86-AS1 |
LY86 antisense RNA 1 |
105584 |
0.07 |
| chr5_14203788_14204073 | 0.26 |
TRIO |
trio Rho guanine nucleotide exchange factor |
20023 |
0.29 |
| chr5_149976234_149976439 | 0.26 |
SYNPO |
synaptopodin |
4306 |
0.19 |
| chr22_38137524_38137675 | 0.26 |
TRIOBP |
TRIO and F-actin binding protein |
4636 |
0.14 |
| chr2_173145900_173146235 | 0.26 |
ENSG00000238572 |
. |
125219 |
0.05 |
| chr12_94657868_94658081 | 0.26 |
PLXNC1 |
plexin C1 |
1677 |
0.34 |
| chr2_25208112_25208263 | 0.26 |
DNAJC27-AS1 |
DNAJC27 antisense RNA 1 |
13135 |
0.13 |
| chr3_53191576_53191739 | 0.26 |
PRKCD |
protein kinase C, delta |
1632 |
0.35 |
| chr6_16682943_16683094 | 0.26 |
RP1-151F17.1 |
|
78351 |
0.11 |
| chr3_66449033_66449184 | 0.26 |
ENSG00000241587 |
. |
74829 |
0.11 |
| chr5_1386620_1386888 | 0.26 |
SLC6A3 |
solute carrier family 6 (neurotransmitter transporter), member 3 |
14668 |
0.2 |
| chr12_110310303_110310470 | 0.25 |
ENSG00000241413 |
. |
6072 |
0.15 |
| chr4_97502612_97502891 | 0.25 |
ENSG00000201640 |
. |
232562 |
0.02 |
| chr1_87615183_87615459 | 0.25 |
ENSG00000221222 |
. |
1660 |
0.51 |
| chr17_54701720_54701923 | 0.25 |
NOG |
noggin |
30761 |
0.23 |
| chr14_92730704_92730855 | 0.25 |
ENSG00000201097 |
. |
1267 |
0.55 |
| chr1_17559878_17560367 | 0.25 |
PADI1 |
peptidyl arginine deiminase, type I |
272 |
0.9 |
| chr20_30156414_30156800 | 0.25 |
HM13-AS1 |
HM13 antisense RNA 1 |
4459 |
0.13 |
| chr2_237880972_237881333 | 0.25 |
ENSG00000202341 |
. |
42796 |
0.17 |
| chr8_61862675_61863209 | 0.25 |
CLVS1 |
clavesin 1 |
106775 |
0.07 |
| chr6_38123150_38123303 | 0.25 |
ENSG00000200706 |
. |
51824 |
0.17 |
| chr2_28182385_28182785 | 0.25 |
ENSG00000265321 |
. |
36649 |
0.16 |
| chr1_230262720_230263119 | 0.25 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
59901 |
0.13 |
| chr14_75406536_75406708 | 0.25 |
PGF |
placental growth factor |
15323 |
0.13 |
| chr1_17388211_17388464 | 0.24 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
7672 |
0.18 |
| chr19_10416474_10416677 | 0.24 |
ZGLP1 |
zinc finger, GATA-like protein 1 |
3884 |
0.09 |
| chr13_108520309_108520631 | 0.24 |
FAM155A |
family with sequence similarity 155, member A |
1387 |
0.54 |
| chr1_110776636_110776787 | 0.24 |
KCNC4 |
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
22589 |
0.12 |
| chrX_13506892_13507043 | 0.24 |
EGFL6 |
EGF-like-domain, multiple 6 |
80757 |
0.09 |
| chr6_43806228_43806543 | 0.24 |
VEGFA |
vascular endothelial growth factor A |
64295 |
0.1 |
| chr17_10030593_10030821 | 0.24 |
GAS7 |
growth arrest-specific 7 |
12837 |
0.2 |
| chr2_60610274_60610606 | 0.24 |
ENSG00000200807 |
. |
1300 |
0.5 |
| chr11_1845805_1845956 | 0.24 |
SYT8 |
synaptotagmin VIII |
2829 |
0.14 |
| chr8_35719083_35719234 | 0.24 |
AC012215.1 |
Uncharacterized protein |
69793 |
0.14 |
| chr6_134342442_134342593 | 0.24 |
SLC2A12 |
solute carrier family 2 (facilitated glucose transporter), member 12 |
31257 |
0.19 |
| chr6_151362493_151362728 | 0.24 |
ENSG00000221469 |
. |
1503 |
0.38 |
| chr9_22170998_22171149 | 0.24 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
57396 |
0.15 |
| chr3_123337794_123338198 | 0.24 |
MYLK |
myosin light chain kinase |
1365 |
0.44 |
| chr5_138363011_138363258 | 0.24 |
CTB-46B19.2 |
|
14795 |
0.16 |
| chr12_48001731_48001929 | 0.23 |
RPAP3 |
RNA polymerase II associated protein 3 |
97939 |
0.07 |
| chr14_77249034_77249359 | 0.23 |
RP11-488C13.6 |
|
1335 |
0.4 |
| chr1_45292295_45292814 | 0.23 |
PTCH2 |
patched 2 |
3622 |
0.11 |
| chr4_141003180_141003333 | 0.23 |
RP11-392B6.1 |
|
45913 |
0.17 |
| chr13_61146576_61146871 | 0.23 |
ENSG00000223076 |
. |
41019 |
0.2 |
| chr10_10563093_10563244 | 0.23 |
ENSG00000200849 |
. |
288594 |
0.01 |
| chr21_16624814_16624965 | 0.23 |
NRIP1 |
nuclear receptor interacting protein 1 |
187568 |
0.03 |
| chr8_145078675_145078826 | 0.23 |
SPATC1 |
spermatogenesis and centriole associated 1 |
7832 |
0.09 |
| chr2_105941174_105941337 | 0.23 |
TGFBRAP1 |
transforming growth factor, beta receptor associated protein 1 |
5236 |
0.18 |
| chr10_31023932_31024083 | 0.23 |
SVILP1 |
supervillin pseudogene 1 |
39157 |
0.18 |
| chr2_178147224_178147445 | 0.23 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
5171 |
0.12 |
| chr11_47963257_47963527 | 0.23 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
38721 |
0.15 |
| chr2_65084933_65085167 | 0.23 |
ENSG00000199964 |
. |
23759 |
0.15 |
| chr10_3793162_3793446 | 0.23 |
RP11-184A2.3 |
|
45 |
0.98 |
| chrX_13376029_13376315 | 0.23 |
ATXN3L |
ataxin 3-like |
37654 |
0.19 |
| chr2_16817031_16817182 | 0.23 |
FAM49A |
family with sequence similarity 49, member A |
12762 |
0.3 |
| chr1_66955749_66956137 | 0.23 |
SGIP1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
43123 |
0.18 |
| chr21_44994556_44994959 | 0.23 |
HSF2BP |
heat shock transcription factor 2 binding protein |
83268 |
0.08 |
| chr10_31986594_31986822 | 0.23 |
ENSG00000222412 |
. |
58356 |
0.16 |
| chr21_38856192_38856343 | 0.23 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
4820 |
0.27 |
| chr6_116878944_116879095 | 0.23 |
FAM26D |
family with sequence similarity 26, member D |
4106 |
0.16 |
| chr17_42619385_42619639 | 0.23 |
FZD2 |
frizzled family receptor 2 |
15413 |
0.17 |
| chr5_138414055_138414601 | 0.23 |
SIL1 |
SIL1 nucleotide exchange factor |
35957 |
0.15 |
| chr11_119454813_119455176 | 0.23 |
RP11-196E1.3 |
|
24141 |
0.19 |
| chr17_1987997_1988266 | 0.23 |
RP11-667K14.5 |
|
4844 |
0.12 |
| chr14_35854447_35854675 | 0.23 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
18785 |
0.23 |
| chr18_61778130_61778281 | 0.22 |
SERPINB8 |
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
131163 |
0.05 |
| chr18_74211607_74211861 | 0.22 |
RP11-17M16.1 |
uncharacterized protein LOC400658 |
4257 |
0.19 |
| chr3_48597793_48598316 | 0.22 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
508 |
0.63 |
| chr2_123731892_123732043 | 0.22 |
ENSG00000221689 |
. |
707824 |
0.0 |
| chr10_73627738_73628072 | 0.22 |
PSAP |
prosaposin |
16779 |
0.19 |
| chr4_185819150_185819410 | 0.22 |
ENSG00000263458 |
. |
40314 |
0.14 |
| chr10_75827961_75828112 | 0.22 |
VCL |
vinculin |
15198 |
0.2 |
| chr2_235732945_235733096 | 0.22 |
SH3BP4 |
SH3-domain binding protein 4 |
127597 |
0.06 |
| chr15_99366117_99366417 | 0.22 |
ENSG00000264480 |
. |
38612 |
0.17 |
| chr15_67316146_67316685 | 0.22 |
SMAD3 |
SMAD family member 3 |
39686 |
0.2 |
| chr17_36604180_36604649 | 0.22 |
ENSG00000260833 |
. |
3518 |
0.2 |
| chr2_86033426_86033673 | 0.22 |
AC105053.3 |
|
8704 |
0.16 |
| chr6_34302944_34303141 | 0.22 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
57409 |
0.11 |
| chr2_160817652_160817857 | 0.22 |
LY75-CD302 |
LY75-CD302 readthrough |
56533 |
0.13 |
| chr2_226982606_226982904 | 0.22 |
ENSG00000263363 |
. |
540754 |
0.0 |
| chr11_68209017_68209227 | 0.22 |
LRP5 |
low density lipoprotein receptor-related protein 5 |
1781 |
0.36 |
| chr17_58648261_58648412 | 0.22 |
RP11-15E18.4 |
Uncharacterized protein |
6428 |
0.18 |
| chr16_88989864_88990015 | 0.22 |
RP11-830F9.7 |
|
14225 |
0.11 |
| chr20_34761813_34761964 | 0.22 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
197 |
0.93 |
| chr6_18330462_18330649 | 0.22 |
ENSG00000200681 |
. |
23245 |
0.17 |
| chr2_240171631_240171851 | 0.22 |
ENSG00000265215 |
. |
55416 |
0.1 |
| chr12_20647309_20647460 | 0.21 |
RP11-284H19.1 |
|
124188 |
0.06 |
| chr20_58627325_58627485 | 0.21 |
C20orf197 |
chromosome 20 open reading frame 197 |
3575 |
0.3 |
| chr8_145017107_145017820 | 0.21 |
PLEC |
plectin |
663 |
0.52 |
| chr16_85363617_85363768 | 0.21 |
ENSG00000266307 |
. |
23761 |
0.19 |
| chr11_114163796_114163947 | 0.21 |
NNMT |
nicotinamide N-methyltransferase |
2676 |
0.32 |
| chr10_81060018_81060169 | 0.21 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
5882 |
0.26 |
| chr15_101690726_101690920 | 0.21 |
RP11-505E24.2 |
|
64552 |
0.11 |
| chr14_75922531_75922716 | 0.21 |
JDP2 |
Jun dimerization protein 2 |
23786 |
0.17 |
| chr5_73729978_73730173 | 0.21 |
ENSG00000244326 |
. |
116264 |
0.06 |
| chr5_136899227_136899378 | 0.21 |
ENSG00000221612 |
. |
30159 |
0.2 |
| chr11_117959166_117959354 | 0.21 |
TMPRSS4-AS1 |
TMPRSS4 antisense RNA 1 |
1752 |
0.3 |
| chr17_32563089_32563474 | 0.21 |
CCL2 |
chemokine (C-C motif) ligand 2 |
19023 |
0.12 |
| chr2_25805338_25805608 | 0.21 |
DTNB |
dystrobrevin, beta |
67620 |
0.12 |
| chr6_22025591_22025742 | 0.21 |
ENSG00000222515 |
. |
60107 |
0.16 |
| chr2_27302580_27302731 | 0.21 |
EMILIN1 |
elastin microfibril interfacer 1 |
1220 |
0.22 |
| chr10_47659322_47659473 | 0.21 |
ANTXRL |
anthrax toxin receptor-like |
1148 |
0.52 |
| chr14_100882789_100883046 | 0.21 |
RP11-362L22.1 |
|
9675 |
0.13 |
| chr9_92750154_92750359 | 0.21 |
ENSG00000263967 |
. |
35561 |
0.25 |
| chr15_38116485_38116636 | 0.21 |
TMCO5A |
transmembrane and coiled-coil domains 5A |
97580 |
0.09 |
| chr16_85796000_85796186 | 0.21 |
C16orf74 |
chromosome 16 open reading frame 74 |
11358 |
0.1 |
| chr2_74803113_74803707 | 0.21 |
LOXL3 |
lysyl oxidase-like 3 |
20593 |
0.09 |
| chr18_11893414_11893565 | 0.20 |
MPPE1 |
metallophosphoesterase 1 |
3554 |
0.17 |
| chr17_79396946_79397153 | 0.20 |
RP11-1055B8.7 |
BAH and coiled-coil domain-containing protein 1 |
8347 |
0.11 |
| chr1_235093577_235093777 | 0.20 |
ENSG00000239690 |
. |
53744 |
0.15 |
| chr17_77999485_77999773 | 0.20 |
CTD-2529O21.2 |
|
1899 |
0.27 |
| chr2_85128601_85128752 | 0.20 |
TMSB10 |
thymosin beta 10 |
4073 |
0.21 |
| chrX_13269027_13269605 | 0.20 |
GS1-600G8.3 |
|
59455 |
0.14 |
| chr22_25797043_25797363 | 0.20 |
LRP5L |
low density lipoprotein receptor-related protein 5-like |
4141 |
0.23 |
| chr5_73092141_73092406 | 0.20 |
CTC-575I10.1 |
|
9957 |
0.2 |
| chr11_73100197_73100477 | 0.20 |
RP11-809N8.2 |
|
7144 |
0.15 |
| chr11_11438455_11438655 | 0.20 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
63651 |
0.12 |
| chr10_111650492_111650643 | 0.20 |
XPNPEP1 |
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
17010 |
0.2 |
| chr5_66000661_66000812 | 0.20 |
MAST4-IT1 |
MAST4 intronic transcript 1 (non-protein coding) |
42577 |
0.19 |
| chr17_40063221_40063372 | 0.20 |
ACLY |
ATP citrate lyase |
11908 |
0.12 |
| chr3_24283691_24283842 | 0.20 |
THRB |
thyroid hormone receptor, beta |
76680 |
0.12 |
| chr11_123175979_123176130 | 0.20 |
ENSG00000200496 |
. |
3956 |
0.28 |
| chr20_33159345_33159531 | 0.20 |
MAP1LC3A |
microtubule-associated protein 1 light chain 3 alpha |
12916 |
0.17 |
| chr3_47051591_47051826 | 0.20 |
NBEAL2 |
neurobeachin-like 2 |
4937 |
0.21 |
| chr1_16500642_16500847 | 0.20 |
RP11-276H7.3 |
|
14983 |
0.1 |
| chr1_230281309_230281781 | 0.20 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
78527 |
0.1 |
| chr21_30688780_30689338 | 0.20 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
10391 |
0.18 |
| chr11_67124730_67125439 | 0.20 |
POLD4 |
polymerase (DNA-directed), delta 4, accessory subunit |
4067 |
0.1 |
| chr1_178027486_178027784 | 0.20 |
RASAL2 |
RAS protein activator like 2 |
35229 |
0.22 |
| chr20_49306472_49306623 | 0.20 |
FAM65C |
family with sequence similarity 65, member C |
1369 |
0.41 |
| chr10_104389775_104390433 | 0.20 |
TRIM8 |
tripartite motif containing 8 |
14149 |
0.16 |
| chr8_97872343_97872494 | 0.20 |
CPQ |
carboxypeptidase Q |
98950 |
0.09 |
| chr3_99129524_99129675 | 0.20 |
ENSG00000266030 |
. |
43866 |
0.2 |
| chr18_42828323_42828591 | 0.20 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
35497 |
0.2 |
| chr9_128289880_128290161 | 0.20 |
ENSG00000243845 |
. |
14534 |
0.21 |
| chr2_36603812_36603963 | 0.20 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
20273 |
0.27 |
| chr8_102388155_102388449 | 0.19 |
ENSG00000239211 |
. |
21859 |
0.22 |
| chr6_11324269_11324660 | 0.19 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
58068 |
0.14 |
| chr13_72406066_72406303 | 0.19 |
DACH1 |
dachshund homolog 1 (Drosophila) |
34723 |
0.24 |
| chr2_65581059_65581210 | 0.19 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
9214 |
0.23 |
| chr14_101131713_101131864 | 0.19 |
DLK1 |
delta-like 1 homolog (Drosophila) |
60254 |
0.06 |
| chr17_56386632_56386867 | 0.19 |
BZRAP1 |
benzodiazepine receptor (peripheral) associated protein 1 |
825 |
0.48 |
| chr3_11243097_11243459 | 0.19 |
HRH1 |
histamine receptor H1 |
24439 |
0.24 |
| chr13_114822172_114822499 | 0.19 |
RASA3 |
RAS p21 protein activator 3 |
21103 |
0.23 |
| chr13_80934002_80934271 | 0.19 |
ENSG00000202398 |
. |
9533 |
0.26 |
| chr22_18042634_18042900 | 0.19 |
SLC25A18 |
solute carrier family 25 (glutamate carrier), member 18 |
372 |
0.87 |
| chr1_192800189_192800340 | 0.19 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
22093 |
0.22 |
| chr8_142156822_142157077 | 0.19 |
DENND3 |
DENN/MADD domain containing 3 |
5647 |
0.21 |
| chr11_11990885_11991036 | 0.19 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
39170 |
0.2 |
| chr17_56584843_56584994 | 0.19 |
MTMR4 |
myotubularin related protein 4 |
6479 |
0.13 |
| chr18_9838569_9838743 | 0.19 |
ENSG00000252680 |
. |
6187 |
0.19 |
| chr17_57863390_57863857 | 0.19 |
RP11-619I22.1 |
|
1449 |
0.37 |
| chr16_11373189_11373514 | 0.19 |
PRM1 |
protamine 1 |
1856 |
0.15 |
| chr5_157295124_157295340 | 0.19 |
CLINT1 |
clathrin interactor 1 |
9049 |
0.23 |
| chr10_104953660_104953838 | 0.19 |
NT5C2 |
5'-nucleotidase, cytosolic II |
693 |
0.72 |
| chr16_55542103_55542255 | 0.19 |
LPCAT2 |
lysophosphatidylcholine acyltransferase 2 |
731 |
0.71 |
| chr9_132096828_132097338 | 0.19 |
ENSG00000242281 |
. |
35657 |
0.16 |
| chr1_73100855_73101006 | 0.19 |
ENSG00000212366 |
. |
82522 |
0.12 |
| chr11_36488512_36488753 | 0.19 |
RP11-219O3.2 |
|
40960 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.2 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 0.2 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0060087 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0032351 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.2 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0051852 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.2 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0014052 | gamma-aminobutyric acid secretion(GO:0014051) regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0003309 | type B pancreatic cell differentiation(GO:0003309) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.0 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.2 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.0 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0071071 | regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0006896 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0010894 | negative regulation of steroid biosynthetic process(GO:0010894) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0071295 | cellular response to vitamin(GO:0071295) |
| 0.0 | 0.0 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.1 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.0 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.0 | GO:0046881 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.0 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.0 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.0 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.0 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.1 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0031083 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.0 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.0 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0000036 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |