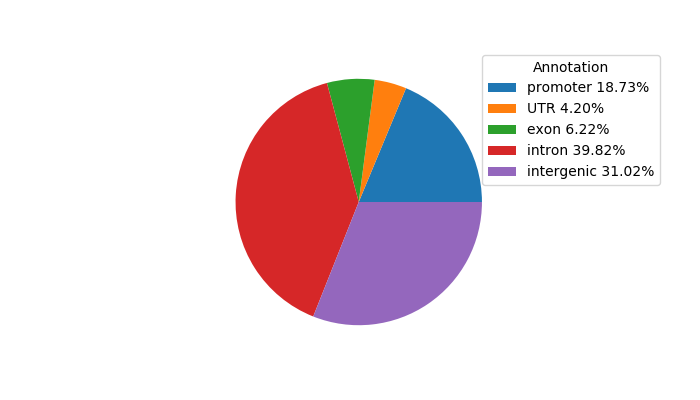
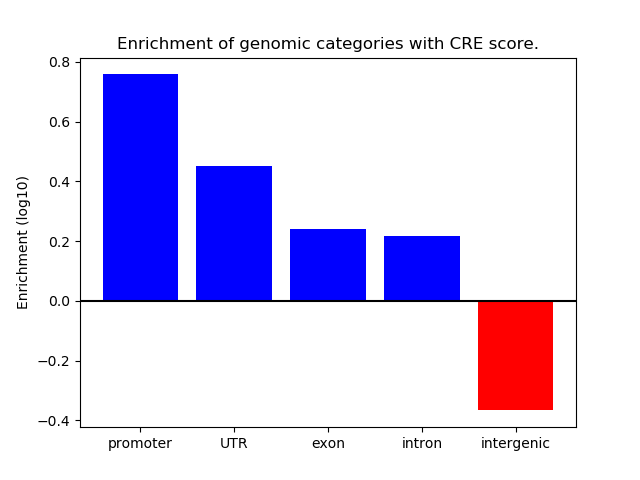
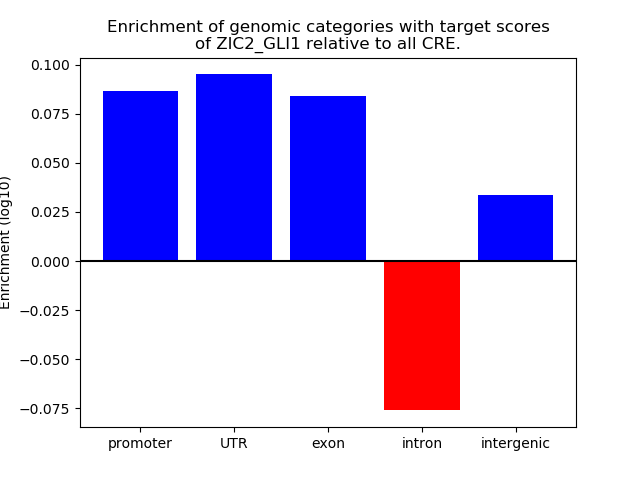
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for ZIC2_GLI1
Z-value: 0.59
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
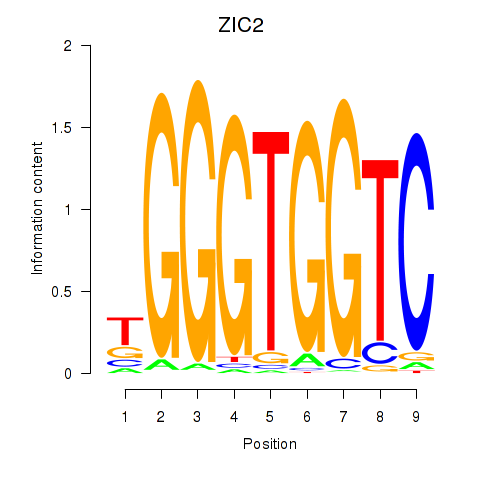
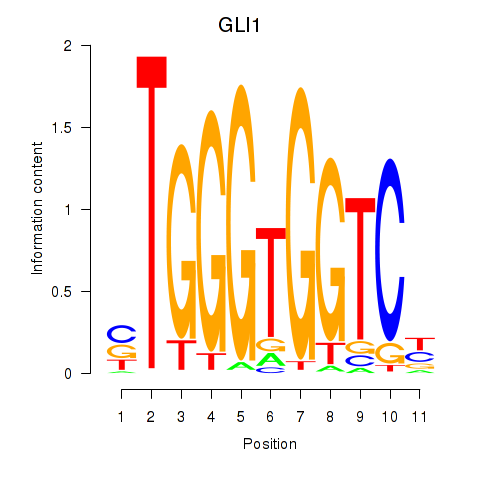
ZIC2
|
ENSG00000043355.6 | Zic family member 2 |
|
GLI1
|
ENSG00000111087.5 | GLI family zinc finger 1 |
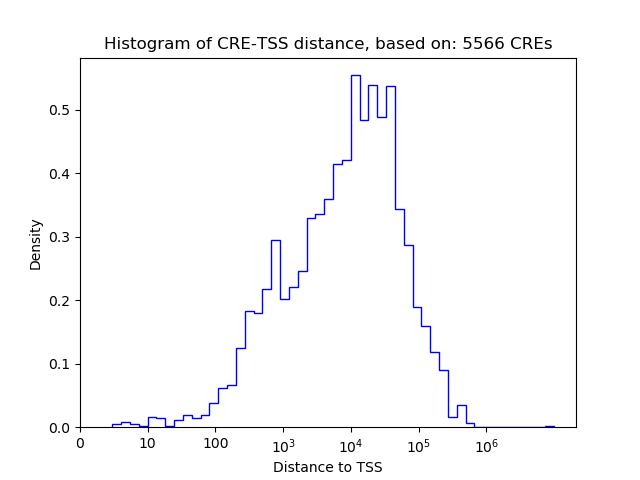
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_57854196_57854445 | GLI1 | 46 | 0.938437 | -0.59 | 9.4e-02 | Click! |
| chr12_57855026_57855259 | GLI1 | 868 | 0.360981 | -0.49 | 1.8e-01 | Click! |
| chr12_57855717_57856166 | GLI1 | 1534 | 0.193930 | -0.40 | 2.9e-01 | Click! |
| chr12_57853153_57853708 | GLI1 | 488 | 0.601516 | -0.38 | 3.2e-01 | Click! |
| chr12_57855309_57855460 | GLI1 | 1110 | 0.275992 | -0.34 | 3.7e-01 | Click! |
| chr13_100632410_100632783 | ZIC2 | 1430 | 0.406983 | -0.80 | 8.9e-03 | Click! |
| chr13_100634648_100635004 | ZIC2 | 800 | 0.627942 | -0.68 | 4.6e-02 | Click! |
| chr13_100635035_100635186 | ZIC2 | 1084 | 0.507643 | -0.67 | 4.6e-02 | Click! |
| chr13_100631814_100632089 | ZIC2 | 2075 | 0.305579 | -0.66 | 5.4e-02 | Click! |
| chr13_100633451_100633876 | ZIC2 | 363 | 0.867073 | -0.64 | 6.3e-02 | Click! |
Activity of the ZIC2_GLI1 motif across conditions
Conditions sorted by the z-value of the ZIC2_GLI1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
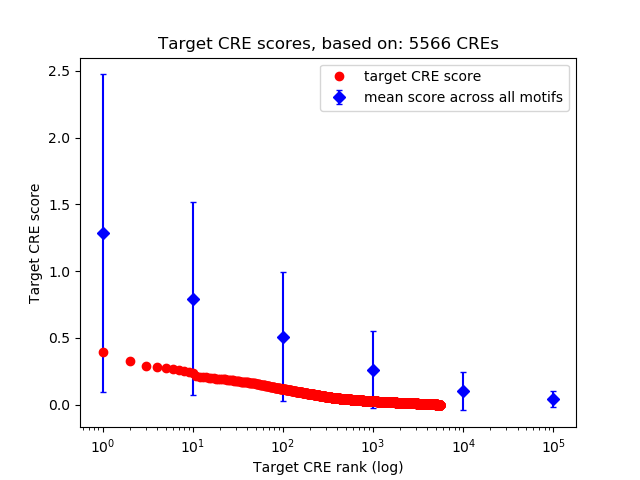
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr21_39617669_39617820 | 0.39 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
10919 |
0.26 |
| chr6_52475238_52475389 | 0.32 |
TRAM2 |
translocation associated membrane protein 2 |
33600 |
0.17 |
| chr3_27576510_27576661 | 0.29 |
ENSG00000238912 |
. |
12764 |
0.2 |
| chr1_95257800_95258262 | 0.28 |
SLC44A3 |
solute carrier family 44, member 3 |
27867 |
0.18 |
| chr9_36625036_36625242 | 0.28 |
MELK |
maternal embryonic leucine zipper kinase |
52231 |
0.16 |
| chr6_37093849_37094027 | 0.27 |
PIM1 |
pim-1 oncogene |
44041 |
0.13 |
| chr17_79057073_79057372 | 0.26 |
BAIAP2 |
BAI1-associated protein 2 |
3039 |
0.17 |
| chr6_16801385_16801811 | 0.25 |
RP1-151F17.1 |
|
39455 |
0.18 |
| chr8_123527926_123528077 | 0.24 |
ENSG00000238901 |
. |
155529 |
0.04 |
| chr5_150400042_150400772 | 0.24 |
GPX3 |
glutathione peroxidase 3 (plasma) |
240 |
0.93 |
| chr14_73706789_73707006 | 0.22 |
PAPLN |
papilin, proteoglycan-like sulfated glycoprotein |
564 |
0.69 |
| chr2_12013931_12014082 | 0.21 |
ENSG00000265172 |
. |
36894 |
0.17 |
| chr11_69795112_69795263 | 0.21 |
ANO1-AS2 |
ANO1 antisense RNA 2 (head to head) |
126267 |
0.04 |
| chr16_88839995_88840537 | 0.20 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
11353 |
0.09 |
| chr5_179341315_179341544 | 0.20 |
TBC1D9B |
TBC1 domain family, member 9B (with GRAM domain) |
6570 |
0.16 |
| chr18_52820659_52820810 | 0.20 |
ENSG00000252437 |
. |
6858 |
0.3 |
| chr2_173991432_173991997 | 0.20 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
36367 |
0.18 |
| chr1_38587427_38587578 | 0.20 |
ENSG00000265596 |
. |
32599 |
0.17 |
| chr1_41965874_41966025 | 0.19 |
EDN2 |
endothelin 2 |
15607 |
0.19 |
| chr12_120933579_120933730 | 0.19 |
DYNLL1-AS1 |
DYNLL1 antisense RNA 1 |
89 |
0.65 |
| chr14_70227367_70227518 | 0.19 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
6368 |
0.23 |
| chr12_4773711_4773869 | 0.19 |
NDUFA9 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
11123 |
0.15 |
| chr14_103006186_103006337 | 0.19 |
ENSG00000266015 |
. |
280 |
0.9 |
| chr17_10100992_10101234 | 0.19 |
GAS7 |
growth arrest-specific 7 |
755 |
0.71 |
| chr1_858767_859188 | 0.19 |
SAMD11 |
sterile alpha motif domain containing 11 |
1283 |
0.3 |
| chr6_163148998_163149579 | 0.18 |
PACRG |
PARK2 co-regulated |
295 |
0.67 |
| chr15_36654222_36654373 | 0.18 |
C15orf41 |
chromosome 15 open reading frame 41 |
217515 |
0.02 |
| chr1_220863452_220864325 | 0.18 |
C1orf115 |
chromosome 1 open reading frame 115 |
701 |
0.71 |
| chr3_11391565_11391730 | 0.18 |
ATG7 |
autophagy related 7 |
14484 |
0.28 |
| chr8_8503148_8503299 | 0.18 |
ENSG00000264445 |
. |
12249 |
0.23 |
| chr16_29836315_29836998 | 0.18 |
MVP |
major vault protein |
4010 |
0.08 |
| chr18_24022178_24022329 | 0.17 |
ENSG00000207160 |
. |
75883 |
0.1 |
| chr19_35496336_35496487 | 0.17 |
GRAMD1A |
GRAM domain containing 1A |
5051 |
0.11 |
| chr19_1017328_1017803 | 0.17 |
TMEM259 |
transmembrane protein 259 |
143 |
0.89 |
| chr4_157852774_157852925 | 0.17 |
PDGFC |
platelet derived growth factor C |
39206 |
0.17 |
| chr11_65682854_65683231 | 0.17 |
C11orf68 |
chromosome 11 open reading frame 68 |
2789 |
0.12 |
| chr1_22209366_22209608 | 0.17 |
HSPG2 |
heparan sulfate proteoglycan 2 |
5727 |
0.21 |
| chr11_35953574_35953793 | 0.17 |
LDLRAD3 |
low density lipoprotein receptor class A domain containing 3 |
11848 |
0.18 |
| chr1_205115054_205115205 | 0.17 |
RBBP5 |
retinoblastoma binding protein 5 |
23986 |
0.16 |
| chr10_126830022_126830361 | 0.17 |
CTBP2 |
C-terminal binding protein 2 |
17094 |
0.26 |
| chr1_38945946_38946097 | 0.17 |
ENSG00000200796 |
. |
83690 |
0.1 |
| chr20_43037764_43037915 | 0.16 |
ENSG00000266151 |
. |
1079 |
0.33 |
| chr4_152405646_152405797 | 0.16 |
FAM160A1 |
family with sequence similarity 160, member A1 |
1960 |
0.42 |
| chr2_118883182_118883333 | 0.16 |
INSIG2 |
insulin induced gene 2 |
37207 |
0.19 |
| chr3_188994860_188995296 | 0.16 |
TPRG1-AS2 |
TPRG1 antisense RNA 2 |
36695 |
0.21 |
| chr9_138947554_138948458 | 0.16 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
5580 |
0.24 |
| chrX_133804180_133804468 | 0.16 |
PLAC1 |
placenta-specific 1 |
11811 |
0.21 |
| chr17_80192804_80193076 | 0.16 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
11 |
0.96 |
| chr3_71864795_71864946 | 0.16 |
ENSG00000239250 |
. |
11458 |
0.21 |
| chr4_84285334_84285485 | 0.16 |
HPSE |
heparanase |
29103 |
0.19 |
| chr5_78051704_78051855 | 0.16 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
107131 |
0.07 |
| chr6_131323866_131324017 | 0.15 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
2034 |
0.47 |
| chr6_150623249_150623400 | 0.15 |
ENSG00000201628 |
. |
24575 |
0.21 |
| chr17_1971701_1971993 | 0.15 |
HIC1 |
hypermethylated in cancer 1 |
12243 |
0.09 |
| chr3_37932792_37933048 | 0.15 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
29255 |
0.15 |
| chr10_96034565_96034716 | 0.15 |
PLCE1-AS1 |
PLCE1 antisense RNA 1 |
12164 |
0.19 |
| chr12_125550229_125550908 | 0.15 |
AACS |
acetoacetyl-CoA synthetase |
581 |
0.82 |
| chr16_85160354_85160505 | 0.15 |
FAM92B |
family with sequence similarity 92, member B |
14315 |
0.21 |
| chr7_130131394_130131755 | 0.15 |
MEST |
mesoderm specific transcript |
360 |
0.78 |
| chr5_37722434_37722606 | 0.15 |
ENSG00000206743 |
. |
21981 |
0.23 |
| chr6_35461097_35461248 | 0.14 |
TEAD3 |
TEA domain family member 3 |
3555 |
0.21 |
| chr19_732760_732911 | 0.14 |
MISP |
mitotic spindle positioning |
18291 |
0.09 |
| chr14_88217881_88218032 | 0.14 |
GALC |
galactosylceramidase |
241226 |
0.02 |
| chr2_97513815_97513966 | 0.14 |
ANKRD23 |
ankyrin repeat domain 23 |
4119 |
0.15 |
| chr22_44555485_44555636 | 0.14 |
PARVG |
parvin, gamma |
13276 |
0.26 |
| chr12_50190526_50190677 | 0.14 |
NCKAP5L |
NCK-associated protein 5-like |
184 |
0.94 |
| chr4_82177498_82177708 | 0.14 |
PRKG2 |
protein kinase, cGMP-dependent, type II |
41385 |
0.18 |
| chr5_171057316_171057470 | 0.14 |
ENSG00000264303 |
. |
140141 |
0.04 |
| chrX_143217715_143217866 | 0.14 |
UBE2NL |
ubiquitin-conjugating enzyme E2N-like |
250617 |
0.02 |
| chr7_2874138_2874289 | 0.14 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
9437 |
0.26 |
| chr5_146783504_146783765 | 0.14 |
CTB-108O6.2 |
|
2311 |
0.31 |
| chr16_54966944_54967095 | 0.13 |
IRX5 |
iroquois homeobox 5 |
1035 |
0.7 |
| chr7_129305011_129305162 | 0.13 |
NRF1 |
nuclear respiratory factor 1 |
7895 |
0.23 |
| chr12_9472275_9472568 | 0.13 |
ENSG00000212440 |
. |
33003 |
0.16 |
| chr2_144713214_144713365 | 0.13 |
AC016910.1 |
|
18649 |
0.25 |
| chr12_66393056_66393407 | 0.13 |
RP11-366L20.3 |
|
50972 |
0.12 |
| chr12_26427261_26427559 | 0.13 |
RP11-283G6.5 |
|
2557 |
0.31 |
| chr14_75920905_75921056 | 0.13 |
JDP2 |
Jun dimerization protein 2 |
22143 |
0.17 |
| chr2_88203324_88203475 | 0.13 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
77928 |
0.09 |
| chr3_53106977_53107358 | 0.13 |
ENSG00000266635 |
. |
20153 |
0.16 |
| chr1_9291920_9292071 | 0.13 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
2839 |
0.28 |
| chr3_185463575_185463764 | 0.13 |
ENSG00000265470 |
. |
22023 |
0.19 |
| chr8_144922026_144922763 | 0.13 |
NRBP2 |
nuclear receptor binding protein 2 |
408 |
0.7 |
| chr10_104221677_104221977 | 0.13 |
TMEM180 |
transmembrane protein 180 |
657 |
0.51 |
| chr15_64572463_64572614 | 0.13 |
ENSG00000252774 |
. |
62200 |
0.09 |
| chr1_21916647_21916859 | 0.12 |
ENSG00000221781 |
. |
2288 |
0.25 |
| chr1_202825022_202825173 | 0.12 |
RP11-480I12.5 |
|
3130 |
0.17 |
| chr3_171865088_171865375 | 0.12 |
FNDC3B |
fibronectin type III domain containing 3B |
20467 |
0.25 |
| chr20_1852587_1852738 | 0.12 |
SIRPA |
signal-regulatory protein alpha |
22492 |
0.22 |
| chr16_68368214_68368400 | 0.12 |
ENSG00000222177 |
. |
4875 |
0.11 |
| chr2_237653499_237653650 | 0.12 |
ACKR3 |
atypical chemokine receptor 3 |
175290 |
0.03 |
| chr17_40834813_40835091 | 0.12 |
CCR10 |
chemokine (C-C motif) receptor 10 |
130 |
0.71 |
| chr2_47564059_47564230 | 0.12 |
EPCAM |
epithelial cell adhesion molecule |
8153 |
0.19 |
| chr9_36859369_36859520 | 0.12 |
RP11-344B23.2 |
|
2892 |
0.23 |
| chr6_1821074_1821225 | 0.12 |
FOXC1 |
forkhead box C1 |
210468 |
0.02 |
| chr2_172974903_172975054 | 0.12 |
DLX2 |
distal-less homeobox 2 |
7350 |
0.21 |
| chr4_56212156_56212324 | 0.12 |
SRD5A3 |
steroid 5 alpha-reductase 3 |
36 |
0.98 |
| chr16_73102653_73102971 | 0.12 |
ZFHX3 |
zinc finger homeobox 3 |
9215 |
0.24 |
| chrX_140270368_140270761 | 0.12 |
LDOC1 |
leucine zipper, down-regulated in cancer 1 |
746 |
0.65 |
| chr17_14651949_14652297 | 0.12 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
447723 |
0.01 |
| chr2_120018753_120018963 | 0.12 |
STEAP3-AS1 |
STEAP3 antisense RNA 1 |
12211 |
0.19 |
| chr19_56135251_56135453 | 0.12 |
ZNF784 |
zinc finger protein 784 |
580 |
0.48 |
| chr2_103511305_103511456 | 0.12 |
TMEM182 |
transmembrane protein 182 |
132908 |
0.05 |
| chr2_238624172_238624755 | 0.12 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
23481 |
0.2 |
| chr15_38117070_38117334 | 0.12 |
TMCO5A |
transmembrane and coiled-coil domains 5A |
96938 |
0.09 |
| chr16_2302179_2302431 | 0.11 |
ECI1 |
enoyl-CoA delta isomerase 1 |
4 |
0.91 |
| chr16_85946040_85946191 | 0.11 |
IRF8 |
interferon regulatory factor 8 |
1465 |
0.49 |
| chr14_25591053_25591204 | 0.11 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
71625 |
0.13 |
| chr14_93527679_93527830 | 0.11 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
4949 |
0.22 |
| chr5_96001162_96001313 | 0.11 |
CAST |
calpastatin |
2480 |
0.28 |
| chr2_64699079_64699230 | 0.11 |
LGALSL |
lectin, galactoside-binding-like |
17478 |
0.18 |
| chr10_88730076_88730664 | 0.11 |
RP11-96C23.15 |
|
213 |
0.74 |
| chr9_71180478_71180629 | 0.11 |
TMEM252 |
transmembrane protein 252 |
24770 |
0.23 |
| chr14_69394581_69394867 | 0.11 |
ACTN1 |
actinin, alpha 1 |
19541 |
0.21 |
| chr1_203032220_203032452 | 0.11 |
PPFIA4 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
9462 |
0.13 |
| chr9_137262651_137262802 | 0.11 |
ENSG00000263897 |
. |
8531 |
0.25 |
| chr20_30194564_30194715 | 0.11 |
ENSG00000264395 |
. |
350 |
0.77 |
| chr1_229228845_229228996 | 0.11 |
RP5-1061H20.5 |
|
134389 |
0.04 |
| chr9_35604415_35605122 | 0.11 |
TESK1 |
testis-specific kinase 1 |
599 |
0.54 |
| chr14_74968488_74968639 | 0.11 |
NPC2 |
Niemann-Pick disease, type C2 |
7683 |
0.15 |
| chr20_10589730_10589881 | 0.11 |
JAG1 |
jagged 1 |
53349 |
0.15 |
| chr19_43860021_43860172 | 0.11 |
CD177 |
CD177 molecule |
2271 |
0.26 |
| chr3_49590959_49591457 | 0.11 |
BSN |
bassoon presynaptic cytomatrix protein |
714 |
0.65 |
| chr22_43301928_43302097 | 0.10 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
39867 |
0.15 |
| chr1_44359454_44359605 | 0.10 |
ST3GAL3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
5731 |
0.17 |
| chr10_102288134_102289261 | 0.10 |
HIF1AN |
hypoxia inducible factor 1, alpha subunit inhibitor |
132 |
0.88 |
| chr16_87715628_87715779 | 0.10 |
AC010536.1 |
Uncharacterized protein; cDNA FLJ45526 fis, clone BRTHA2027227 |
14050 |
0.17 |
| chr1_97030873_97031024 | 0.10 |
ENSG00000241992 |
. |
17817 |
0.28 |
| chr10_133999367_134000256 | 0.10 |
RP11-140A10.3 |
|
345 |
0.67 |
| chr22_31502446_31502597 | 0.10 |
SELM |
Selenoprotein M |
1033 |
0.37 |
| chrX_134185283_134186016 | 0.10 |
FAM127B |
family with sequence similarity 127, member B |
556 |
0.74 |
| chr5_97550623_97550774 | 0.10 |
ENSG00000223053 |
. |
424237 |
0.01 |
| chr9_89591706_89591857 | 0.10 |
GAS1 |
growth arrest-specific 1 |
29677 |
0.25 |
| chr14_103739694_103740051 | 0.10 |
RP11-45P15.4 |
|
58832 |
0.09 |
| chr10_5617233_5617384 | 0.10 |
CALML3-AS1 |
CALML3 antisense RNA 1 |
49099 |
0.09 |
| chr7_8092160_8092311 | 0.10 |
AC006042.6 |
|
61420 |
0.11 |
| chr1_3285947_3286098 | 0.10 |
PRDM16 |
PR domain containing 16 |
27033 |
0.21 |
| chr13_43859080_43859231 | 0.10 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
76058 |
0.13 |
| chr2_42651162_42651313 | 0.10 |
ENSG00000221300 |
. |
16280 |
0.21 |
| chr16_3293109_3293260 | 0.10 |
ZNF200 |
zinc finger protein 200 |
6963 |
0.1 |
| chr10_60514271_60514422 | 0.10 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
38949 |
0.22 |
| chr13_114540098_114540530 | 0.10 |
GAS6 |
growth arrest-specific 6 |
1297 |
0.5 |
| chr16_57034102_57034447 | 0.10 |
NLRC5 |
NLR family, CARD domain containing 5 |
3491 |
0.17 |
| chr17_17881945_17882129 | 0.10 |
LRRC48 |
leucine rich repeat containing 48 |
5761 |
0.15 |
| chr10_118892258_118892511 | 0.10 |
VAX1 |
ventral anterior homeobox 1 |
5183 |
0.19 |
| chr2_64899791_64900044 | 0.10 |
SERTAD2 |
SERTA domain containing 2 |
18870 |
0.22 |
| chr1_17520298_17520449 | 0.10 |
PADI1 |
peptidyl arginine deiminase, type I |
11248 |
0.17 |
| chr9_95873040_95873199 | 0.10 |
C9orf89 |
chromosome 9 open reading frame 89 |
14619 |
0.15 |
| chr5_88340795_88340946 | 0.10 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
79178 |
0.12 |
| chr5_36693133_36693338 | 0.10 |
CTD-2353F22.1 |
|
32040 |
0.22 |
| chr1_118644337_118644488 | 0.10 |
SPAG17 |
sperm associated antigen 17 |
83434 |
0.1 |
| chr6_35393359_35394053 | 0.10 |
FANCE |
Fanconi anemia, complementation group E |
26432 |
0.15 |
| chr6_123031592_123031771 | 0.09 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
7025 |
0.24 |
| chr19_45959219_45959447 | 0.09 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
5350 |
0.12 |
| chr20_44638017_44638168 | 0.09 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
545 |
0.67 |
| chr5_147781123_147781294 | 0.09 |
FBXO38 |
F-box protein 38 |
6933 |
0.18 |
| chr11_126216427_126216820 | 0.09 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
8932 |
0.14 |
| chr15_57109611_57109762 | 0.09 |
ZNF280D |
zinc finger protein 280D |
83402 |
0.1 |
| chr2_201535042_201535193 | 0.09 |
AOX1 |
aldehyde oxidase 1 |
7487 |
0.23 |
| chr15_80706158_80706309 | 0.09 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
9540 |
0.19 |
| chr12_11733293_11733560 | 0.09 |
ENSG00000251747 |
. |
34013 |
0.16 |
| chr5_34023864_34024015 | 0.09 |
AMACR |
alpha-methylacyl-CoA racemase |
15719 |
0.18 |
| chr11_70073277_70073428 | 0.09 |
RP11-805J14.5 |
|
19856 |
0.13 |
| chr15_90936797_90937093 | 0.09 |
RP11-154B12.3 |
|
4168 |
0.17 |
| chr1_21194615_21194766 | 0.09 |
ENSG00000251914 |
. |
26956 |
0.16 |
| chr2_225085875_225086167 | 0.09 |
ENSG00000211987 |
. |
18252 |
0.28 |
| chr18_56554770_56555263 | 0.09 |
ZNF532 |
zinc finger protein 532 |
22230 |
0.17 |
| chr13_46076174_46076325 | 0.09 |
COG3 |
component of oligomeric golgi complex 3 |
37189 |
0.14 |
| chr12_40552597_40552758 | 0.09 |
LRRK2 |
leucine-rich repeat kinase 2 |
37869 |
0.17 |
| chr17_44841951_44842102 | 0.09 |
NSF |
N-ethylmaleimide-sensitive factor |
38104 |
0.15 |
| chr16_66440801_66441158 | 0.09 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
11621 |
0.17 |
| chr4_26240278_26240429 | 0.09 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
33876 |
0.24 |
| chr5_43772686_43772978 | 0.09 |
NNT |
nicotinamide nucleotide transhydrogenase |
127373 |
0.05 |
| chr9_107878597_107878922 | 0.09 |
ENSG00000201583 |
. |
19722 |
0.26 |
| chr1_214466045_214466196 | 0.09 |
SMYD2 |
SET and MYND domain containing 2 |
11544 |
0.28 |
| chr1_178252640_178252859 | 0.09 |
RASAL2 |
RAS protein activator like 2 |
57857 |
0.16 |
| chr16_69951449_69951738 | 0.09 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
7300 |
0.16 |
| chr17_79777515_79777666 | 0.09 |
FAM195B |
family with sequence similarity 195, member B |
4773 |
0.08 |
| chr4_48226478_48226629 | 0.09 |
TEC |
tec protein tyrosine kinase |
45328 |
0.14 |
| chr21_37296503_37296654 | 0.09 |
FKSG68 |
|
25851 |
0.19 |
| chr8_134472381_134472532 | 0.09 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
16391 |
0.28 |
| chr7_69119614_69119794 | 0.08 |
AUTS2 |
autism susceptibility candidate 2 |
55107 |
0.16 |
| chr9_140541020_140541171 | 0.08 |
EHMT1 |
euchromatic histone-lysine N-methyltransferase 1 |
27641 |
0.12 |
| chr6_111894573_111894724 | 0.08 |
TRAF3IP2-AS1 |
TRAF3IP2 antisense RNA 1 |
1174 |
0.46 |
| chr8_6086772_6086923 | 0.08 |
RP11-115C21.2 |
|
177216 |
0.03 |
| chr9_137257719_137258146 | 0.08 |
ENSG00000263897 |
. |
13325 |
0.24 |
| chr11_36035998_36036518 | 0.08 |
ENSG00000263389 |
. |
4610 |
0.26 |
| chr20_42378949_42379100 | 0.08 |
GTSF1L |
gametocyte specific factor 1-like |
23386 |
0.19 |
| chr10_122175337_122175488 | 0.08 |
PPAPDC1A |
phosphatidic acid phosphatase type 2 domain containing 1A |
41054 |
0.18 |
| chr7_73869059_73869441 | 0.08 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
811 |
0.66 |
| chr17_48357099_48357487 | 0.08 |
TMEM92 |
transmembrane protein 92 |
5505 |
0.14 |
| chr9_110925898_110926343 | 0.08 |
ENSG00000222512 |
. |
195089 |
0.03 |
| chr3_141679702_141679973 | 0.08 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
2840 |
0.29 |
| chr4_73843280_73843431 | 0.08 |
ENSG00000252955 |
. |
12460 |
0.25 |
| chr9_129135099_129135415 | 0.08 |
MVB12B |
multivesicular body subunit 12B |
3465 |
0.28 |
| chrX_53713767_53714068 | 0.08 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
244 |
0.95 |
| chr1_234791884_234792035 | 0.08 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
46688 |
0.14 |
| chr22_19692388_19692539 | 0.08 |
SEPT5 |
septin 5 |
9524 |
0.18 |
| chr8_143852944_143853095 | 0.08 |
LYNX1 |
Ly6/neurotoxin 1 |
1757 |
0.19 |
| chr8_146077548_146078058 | 0.08 |
COMMD5 |
COMM domain containing 5 |
239 |
0.83 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0003099 | positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |