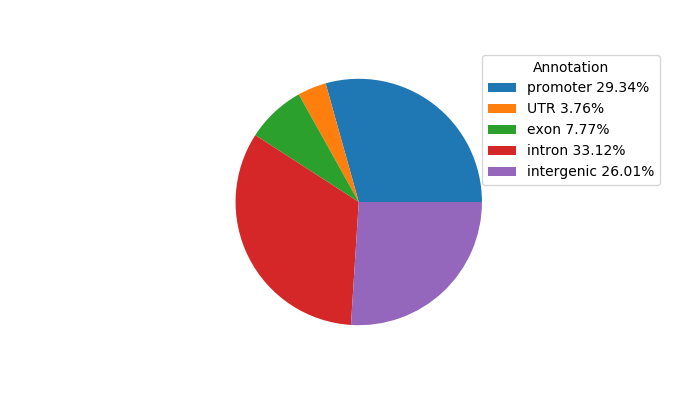
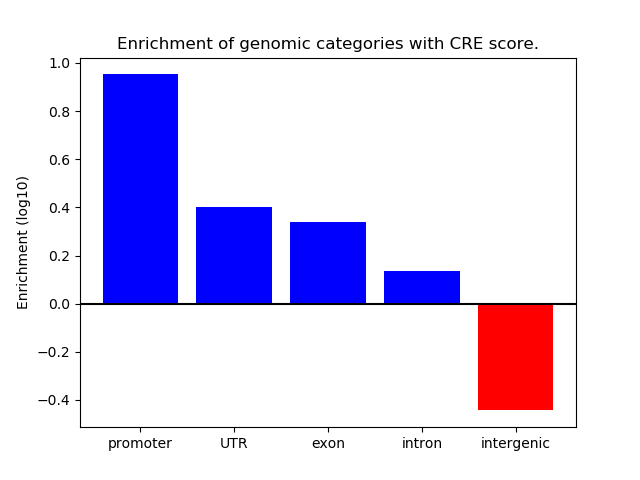
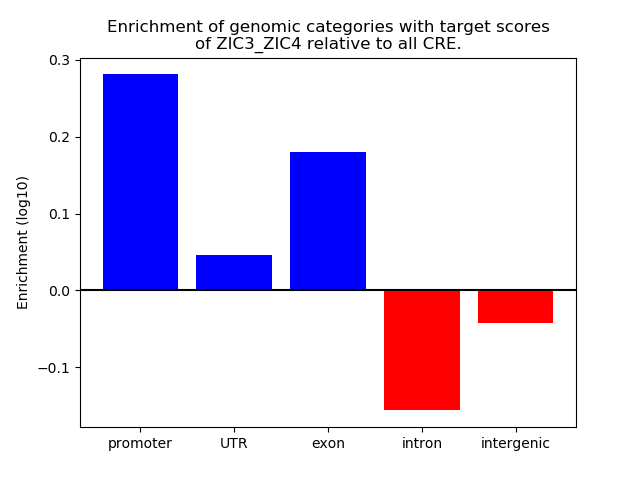
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
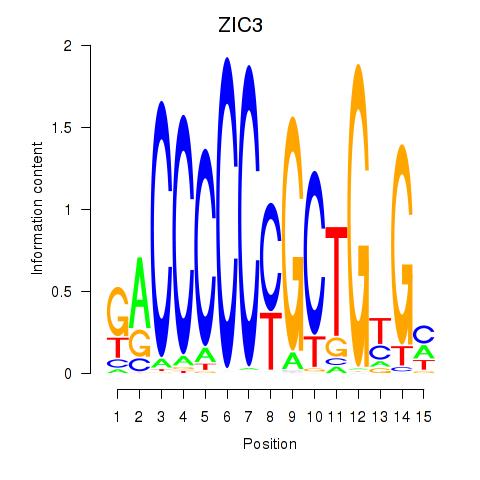
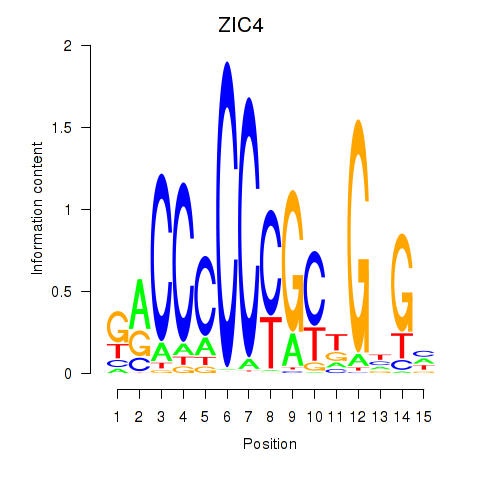
Results for ZIC3_ZIC4
Z-value: 1.65
Transcription factors associated with ZIC3_ZIC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC3
|
ENSG00000156925.7 | Zic family member 3 |
|
ZIC4
|
ENSG00000174963.13 | Zic family member 4 |
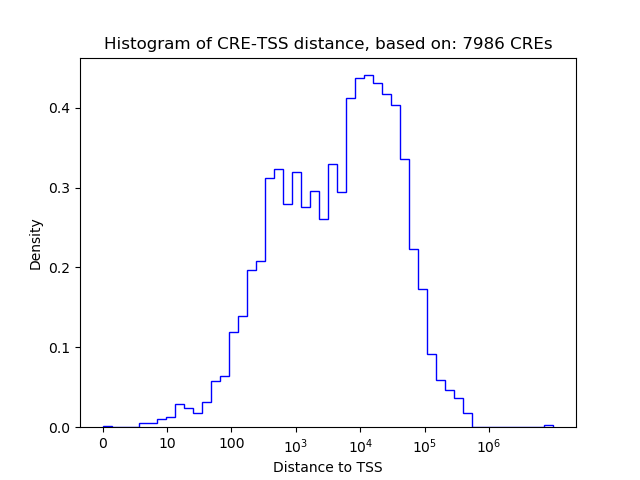
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_136649954_136650105 | ZIC3 | 1352 | 0.440638 | 0.69 | 3.9e-02 | Click! |
| chrX_136647497_136647648 | ZIC3 | 729 | 0.528502 | 0.68 | 4.5e-02 | Click! |
| chrX_136649472_136649623 | ZIC3 | 870 | 0.601861 | 0.58 | 1.0e-01 | Click! |
| chrX_136649682_136649902 | ZIC3 | 1115 | 0.508072 | 0.56 | 1.2e-01 | Click! |
| chrX_136648337_136648602 | ZIC3 | 168 | 0.945410 | 0.56 | 1.2e-01 | Click! |
| chr3_147125253_147125404 | ZIC4 | 681 | 0.634337 | 0.20 | 6.1e-01 | Click! |
Activity of the ZIC3_ZIC4 motif across conditions
Conditions sorted by the z-value of the ZIC3_ZIC4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
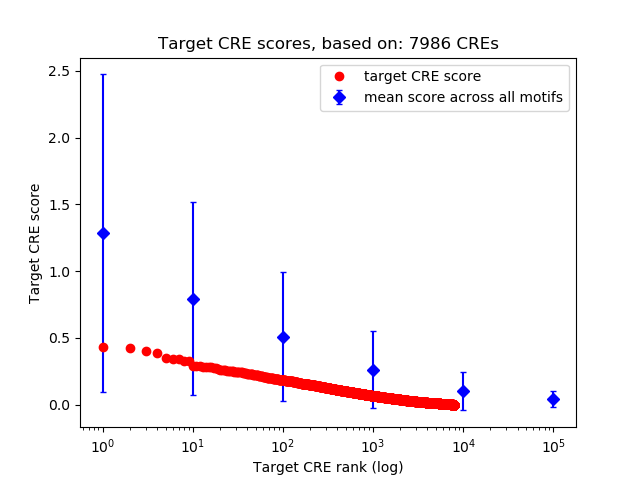
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_38600137_38600500 | 0.43 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
605 |
0.66 |
| chr11_73100197_73100477 | 0.43 |
RP11-809N8.2 |
|
7144 |
0.15 |
| chr17_77904427_77904652 | 0.40 |
TBC1D16 |
TBC1 domain family, member 16 |
20088 |
0.18 |
| chr4_44449502_44449653 | 0.39 |
KCTD8 |
potassium channel tetramerization domain containing 8 |
53 |
0.88 |
| chr3_142840251_142840402 | 0.35 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
2153 |
0.37 |
| chr6_74161154_74161429 | 0.35 |
MB21D1 |
Mab-21 domain containing 1 |
691 |
0.49 |
| chr5_135170958_135171109 | 0.34 |
SLC25A48 |
solute carrier family 25, member 48 |
617 |
0.7 |
| chr8_22977343_22977524 | 0.33 |
TNFRSF10C |
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
16999 |
0.13 |
| chr5_1386620_1386888 | 0.33 |
SLC6A3 |
solute carrier family 6 (neurotransmitter transporter), member 3 |
14668 |
0.2 |
| chr17_72594749_72594939 | 0.29 |
CD300LD |
CD300 molecule-like family member d |
6422 |
0.13 |
| chr10_95315125_95315276 | 0.29 |
FFAR4 |
free fatty acid receptor 4 |
11222 |
0.16 |
| chr20_11241961_11242112 | 0.29 |
C20orf187 |
chromosome 20 open reading frame 187 |
233225 |
0.02 |
| chr1_212872826_212872977 | 0.29 |
BATF3 |
basic leucine zipper transcription factor, ATF-like 3 |
426 |
0.82 |
| chr1_21503657_21503851 | 0.28 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
377 |
0.66 |
| chr9_19551364_19551515 | 0.28 |
RP11-363E7.4 |
|
98232 |
0.07 |
| chr17_35285280_35285503 | 0.28 |
RP11-445F12.1 |
|
8530 |
0.14 |
| chr17_80176111_80176376 | 0.27 |
RP13-516M14.2 |
|
4140 |
0.12 |
| chr9_74384006_74384157 | 0.27 |
TMEM2 |
transmembrane protein 2 |
281 |
0.95 |
| chr7_582328_582615 | 0.27 |
ENSG00000252986 |
. |
7899 |
0.18 |
| chr1_154158713_154158864 | 0.26 |
TPM3 |
tropomyosin 3 |
3104 |
0.13 |
| chr12_111079520_111079671 | 0.26 |
TCTN1 |
tectonic family member 1 |
741 |
0.59 |
| chr17_14822625_14822776 | 0.26 |
ENSG00000238806 |
. |
318500 |
0.01 |
| chr14_76843614_76843766 | 0.26 |
ENSG00000263880 |
. |
22338 |
0.18 |
| chr20_30411382_30411533 | 0.26 |
MYLK2 |
myosin light chain kinase 2 |
4281 |
0.16 |
| chr10_80886709_80887280 | 0.25 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
58202 |
0.14 |
| chr19_4279328_4279696 | 0.25 |
SHD |
Src homology 2 domain containing transforming protein D |
219 |
0.87 |
| chr14_102002023_102002232 | 0.25 |
ENSG00000258498 |
. |
24632 |
0.17 |
| chr17_71586433_71586688 | 0.25 |
RP11-277J6.2 |
|
50605 |
0.13 |
| chr7_45925213_45925504 | 0.25 |
IGFBP1 |
insulin-like growth factor binding protein 1 |
2598 |
0.25 |
| chr18_77331216_77331367 | 0.25 |
AC018445.1 |
Uncharacterized protein |
55234 |
0.15 |
| chr15_70540546_70541057 | 0.25 |
ENSG00000200216 |
. |
55226 |
0.16 |
| chr8_62722273_62722424 | 0.25 |
ENSG00000264408 |
. |
95001 |
0.08 |
| chr6_6728512_6728665 | 0.24 |
LY86-AS1 |
LY86 antisense RNA 1 |
105584 |
0.07 |
| chr17_74025598_74025749 | 0.24 |
EVPL |
envoplakin |
2140 |
0.19 |
| chr10_65224937_65225088 | 0.24 |
JMJD1C-AS1 |
JMJD1C antisense RNA 1 |
23 |
0.93 |
| chr17_37914792_37914943 | 0.24 |
GRB7 |
growth factor receptor-bound protein 7 |
16362 |
0.11 |
| chr12_124875119_124875344 | 0.24 |
NCOR2 |
nuclear receptor corepressor 2 |
1861 |
0.47 |
| chr4_122617648_122617799 | 0.24 |
ANXA5 |
annexin A5 |
394 |
0.89 |
| chr17_74377867_74378161 | 0.24 |
SPHK1 |
sphingosine kinase 1 |
162 |
0.92 |
| chr2_109605860_109606011 | 0.23 |
EDAR |
ectodysplasin A receptor |
107 |
0.98 |
| chr2_66673650_66673801 | 0.23 |
MEIS1 |
Meis homeobox 1 |
3628 |
0.19 |
| chr17_79480328_79480479 | 0.23 |
ACTG1 |
actin, gamma 1 |
503 |
0.46 |
| chr15_90625478_90625629 | 0.23 |
RP11-617F23.1 |
|
114 |
0.95 |
| chr16_17317009_17317184 | 0.23 |
CTD-2576D5.4 |
|
88735 |
0.1 |
| chr16_57831186_57831470 | 0.23 |
KIFC3 |
kinesin family member C3 |
348 |
0.84 |
| chr17_55860787_55861007 | 0.23 |
ENSG00000222976 |
. |
5913 |
0.21 |
| chr13_33412405_33412556 | 0.23 |
ENSG00000207325 |
. |
74790 |
0.11 |
| chr20_60941090_60941476 | 0.23 |
LAMA5 |
laminin, alpha 5 |
1043 |
0.39 |
| chr5_171998769_171998920 | 0.22 |
AC027309.1 |
|
37592 |
0.15 |
| chr16_87871055_87871206 | 0.22 |
RP4-536B24.2 |
|
992 |
0.54 |
| chr1_204959299_204959565 | 0.22 |
NFASC |
neurofascin |
1640 |
0.4 |
| chr3_159579244_159579395 | 0.22 |
SCHIP1 |
schwannomin interacting protein 1 |
8591 |
0.21 |
| chr14_65435464_65435615 | 0.22 |
RAB15 |
RAB15, member RAS oncogene family |
3073 |
0.16 |
| chr2_122014973_122015141 | 0.22 |
TFCP2L1 |
transcription factor CP2-like 1 |
27726 |
0.25 |
| chr5_149867423_149867730 | 0.22 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
1719 |
0.35 |
| chr8_53043724_53043875 | 0.22 |
RP11-26M5.3 |
|
19581 |
0.24 |
| chr14_69122374_69122525 | 0.22 |
CTD-2325P2.4 |
|
27287 |
0.18 |
| chr6_74432173_74432346 | 0.22 |
RP11-553A21.3 |
|
26405 |
0.17 |
| chr22_43506432_43506584 | 0.21 |
BIK |
BCL2-interacting killer (apoptosis-inducing) |
246 |
0.9 |
| chr5_139487909_139488137 | 0.21 |
LINC01024 |
long intergenic non-protein coding RNA 1024 |
127 |
0.95 |
| chr14_96342982_96343133 | 0.21 |
ENSG00000263357 |
. |
25130 |
0.23 |
| chr1_10519294_10519577 | 0.21 |
RP5-1113E3.3 |
|
823 |
0.46 |
| chr15_74325155_74325331 | 0.21 |
PML |
promyelocytic leukemia |
8022 |
0.16 |
| chr21_30688780_30689338 | 0.21 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
10391 |
0.18 |
| chr11_65780370_65780606 | 0.21 |
CST6 |
cystatin E/M |
1176 |
0.27 |
| chr9_38672362_38672574 | 0.20 |
ENSG00000252725 |
. |
7535 |
0.24 |
| chr2_8821447_8821664 | 0.20 |
AC011747.7 |
|
438 |
0.57 |
| chr6_38607899_38608192 | 0.20 |
BTBD9 |
BTB (POZ) domain containing 9 |
121 |
0.97 |
| chr3_17944629_17944780 | 0.20 |
TBC1D5 |
TBC1 domain family, member 5 |
160560 |
0.04 |
| chr11_126216427_126216820 | 0.20 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
8932 |
0.14 |
| chr3_134624316_134624467 | 0.20 |
EPHB1 |
EPH receptor B1 |
53043 |
0.16 |
| chr5_1005247_1005480 | 0.20 |
ENSG00000221244 |
. |
1061 |
0.41 |
| chr7_41852060_41852211 | 0.20 |
AC005027.3 |
|
107202 |
0.07 |
| chr1_114657022_114657208 | 0.20 |
SYT6 |
synaptotagmin VI |
37947 |
0.19 |
| chr1_2158798_2158949 | 0.20 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
1261 |
0.34 |
| chrX_53349853_53350130 | 0.20 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
531 |
0.75 |
| chr16_58497854_58498037 | 0.20 |
NDRG4 |
NDRG family member 4 |
17 |
0.97 |
| chr11_68530734_68530885 | 0.20 |
MTL5 |
metallothionein-like 5, testis-specific (tesmin) |
11777 |
0.2 |
| chr22_27524680_27524831 | 0.20 |
ENSG00000200443 |
. |
92905 |
0.1 |
| chr13_110381697_110381924 | 0.20 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
1181 |
0.62 |
| chr12_52407645_52407796 | 0.20 |
GRASP |
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
2753 |
0.18 |
| chr22_48972497_48972648 | 0.19 |
FAM19A5 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
454 |
0.88 |
| chr2_203025624_203025775 | 0.19 |
AC079354.5 |
|
18415 |
0.15 |
| chr10_30748355_30748506 | 0.19 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
20679 |
0.21 |
| chr2_44058741_44059019 | 0.19 |
ABCG5 |
ATP-binding cassette, sub-family G (WHITE), member 5 |
7009 |
0.18 |
| chr1_10746935_10747086 | 0.19 |
CASZ1 |
castor zinc finger 1 |
41877 |
0.14 |
| chr22_19710642_19710976 | 0.19 |
GP1BB |
glycoprotein Ib (platelet), beta polypeptide |
341 |
0.86 |
| chr5_136827472_136827623 | 0.19 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
6700 |
0.27 |
| chr14_68556146_68556297 | 0.19 |
CTD-2566J3.1 |
|
40692 |
0.18 |
| chr15_31685781_31685934 | 0.19 |
KLF13 |
Kruppel-like factor 13 |
27500 |
0.25 |
| chr16_2198362_2198540 | 0.19 |
RAB26 |
RAB26, member RAS oncogene family |
174 |
0.83 |
| chr1_154378255_154378406 | 0.19 |
IL6R |
interleukin 6 receptor |
239 |
0.79 |
| chr9_1051829_1051980 | 0.19 |
DMRT2 |
doublesex and mab-3 related transcription factor 2 |
290 |
0.93 |
| chr19_13202769_13202920 | 0.19 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
10837 |
0.1 |
| chr13_112547435_112547586 | 0.19 |
ENSG00000264989 |
. |
15638 |
0.29 |
| chr7_23736830_23737011 | 0.18 |
STK31 |
serine/threonine kinase 31 |
12866 |
0.16 |
| chr4_7972689_7972840 | 0.18 |
AFAP1 |
actin filament associated protein 1 |
31111 |
0.14 |
| chr16_89188310_89188461 | 0.18 |
CTD-2555A7.3 |
|
6698 |
0.16 |
| chr10_121080665_121080816 | 0.18 |
RP11-79M19.2 |
|
14954 |
0.2 |
| chr1_54153130_54153281 | 0.18 |
ENSG00000239007 |
. |
1280 |
0.5 |
| chr5_22853160_22853311 | 0.18 |
CDH12 |
cadherin 12, type 2 (N-cadherin 2) |
215 |
0.97 |
| chr1_228352022_228352173 | 0.18 |
IBA57-AS1 |
IBA57 antisense RNA 1 (head to head) |
1116 |
0.29 |
| chr15_91458200_91458390 | 0.18 |
MAN2A2 |
mannosidase, alpha, class 2A, member 2 |
2177 |
0.15 |
| chr6_157882968_157883119 | 0.18 |
ENSG00000266617 |
. |
67121 |
0.12 |
| chr1_204776345_204776581 | 0.18 |
NFASC |
neurofascin |
21319 |
0.2 |
| chr17_48350379_48350530 | 0.18 |
TMEM92 |
transmembrane protein 92 |
1334 |
0.33 |
| chr18_46065806_46065983 | 0.18 |
CTIF |
CBP80/20-dependent translation initiation factor |
108 |
0.98 |
| chr17_80291269_80291420 | 0.18 |
SECTM1 |
secreted and transmembrane 1 |
294 |
0.83 |
| chr16_644060_644236 | 0.18 |
RAB40C |
RAB40C, member RAS oncogene family |
3816 |
0.09 |
| chr9_139716096_139716247 | 0.18 |
RABL6 |
RAB, member RAS oncogene family-like 6 |
390 |
0.64 |
| chr11_20134359_20134761 | 0.18 |
NAV2-AS1 |
NAV2 antisense RNA 1 |
7618 |
0.23 |
| chr4_174139787_174140018 | 0.18 |
RP11-10K16.1 |
|
49099 |
0.11 |
| chr2_100171123_100171274 | 0.18 |
AFF3 |
AF4/FMR2 family, member 3 |
23634 |
0.19 |
| chr20_734961_735112 | 0.18 |
SLC52A3 |
solute carrier family 52 (riboflavin transporter), member 3 |
14095 |
0.2 |
| chrX_46570531_46570682 | 0.18 |
SLC9A7 |
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
47884 |
0.16 |
| chr1_154763833_154763984 | 0.18 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
68411 |
0.08 |
| chr7_150020672_150020823 | 0.18 |
ACTR3C |
ARP3 actin-related protein 3 homolog C (yeast) |
11 |
0.84 |
| chr10_47749182_47749333 | 0.18 |
ANXA8L2 |
annexin A8-like 2 |
2221 |
0.25 |
| chrX_152912384_152912535 | 0.18 |
DUSP9 |
dual specificity phosphatase 9 |
191 |
0.91 |
| chr17_2626915_2627066 | 0.18 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
11033 |
0.11 |
| chr16_3006567_3006722 | 0.18 |
LA16c-321D4.2 |
|
2367 |
0.12 |
| chr3_148184411_148184664 | 0.18 |
AGTR1 |
angiotensin II receptor, type 1 |
231034 |
0.02 |
| chr5_139125701_139125887 | 0.18 |
ENSG00000200756 |
. |
28119 |
0.17 |
| chr5_11384831_11385016 | 0.18 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
204000 |
0.03 |
| chr16_50597873_50598024 | 0.18 |
RP11-401P9.1 |
|
10097 |
0.17 |
| chr22_27812652_27812803 | 0.17 |
RP11-375H17.1 |
|
299741 |
0.01 |
| chr11_119554728_119554879 | 0.17 |
ENSG00000199217 |
. |
27782 |
0.16 |
| chr16_55512713_55512866 | 0.17 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
47 |
0.98 |
| chr2_220435040_220435191 | 0.17 |
OBSL1 |
obscurin-like 1 |
848 |
0.34 |
| chr8_20898687_20898838 | 0.17 |
ENSG00000215945 |
. |
114880 |
0.07 |
| chr5_139063267_139063418 | 0.17 |
CXXC5 |
CXXC finger protein 5 |
4074 |
0.25 |
| chr3_133392987_133393138 | 0.17 |
ENSG00000252641 |
. |
9283 |
0.15 |
| chr5_148645320_148645471 | 0.17 |
AFAP1L1 |
actin filament associated protein 1-like 1 |
6039 |
0.14 |
| chr19_55884009_55884160 | 0.17 |
IL11 |
interleukin 11 |
2253 |
0.11 |
| chr17_68674046_68674197 | 0.17 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
508445 |
0.0 |
| chr21_16236639_16236939 | 0.17 |
AF127577.8 |
|
54066 |
0.15 |
| chr19_39636014_39636168 | 0.17 |
CTC-218B8.3 |
|
8988 |
0.13 |
| chr17_47645178_47645393 | 0.17 |
RP5-1029K10.2 |
|
6141 |
0.13 |
| chr13_22243741_22243916 | 0.17 |
FGF9 |
fibroblast growth factor 9 |
1694 |
0.46 |
| chr21_39812982_39813157 | 0.17 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
57276 |
0.16 |
| chr8_97506971_97507442 | 0.17 |
SDC2 |
syndecan 2 |
970 |
0.67 |
| chr2_46770771_46771016 | 0.17 |
RHOQ |
ras homolog family member Q |
354 |
0.78 |
| chr12_125139086_125139237 | 0.17 |
NCOR2 |
nuclear receptor corepressor 2 |
87151 |
0.1 |
| chr2_114100712_114100863 | 0.17 |
IGKV1OR2-108 |
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
63186 |
0.08 |
| chr8_123378321_123378472 | 0.16 |
ENSG00000238901 |
. |
305134 |
0.01 |
| chr6_158989738_158990048 | 0.16 |
TMEM181 |
transmembrane protein 181 |
32425 |
0.16 |
| chr17_47516412_47516563 | 0.16 |
ENSG00000207127 |
. |
22275 |
0.12 |
| chr1_220498823_220498974 | 0.16 |
RAB3GAP2 |
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
53102 |
0.14 |
| chr15_98447614_98447765 | 0.16 |
ARRDC4 |
arrestin domain containing 4 |
15095 |
0.2 |
| chr20_24763571_24763722 | 0.16 |
CST7 |
cystatin F (leukocystatin) |
166220 |
0.03 |
| chr21_32656703_32656854 | 0.16 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
7695 |
0.31 |
| chr8_41649260_41649615 | 0.16 |
ANK1 |
ankyrin 1, erythrocytic |
5703 |
0.2 |
| chr22_25871447_25871598 | 0.16 |
CTA-390C10.10 |
|
663 |
0.67 |
| chr11_117812434_117812585 | 0.16 |
TMPRSS13 |
transmembrane protease, serine 13 |
12335 |
0.17 |
| chr14_105444909_105445181 | 0.16 |
AHNAK2 |
AHNAK nucleoprotein 2 |
351 |
0.85 |
| chr11_67168630_67168781 | 0.16 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
554 |
0.49 |
| chr10_13701534_13702115 | 0.16 |
RP11-295P9.12 |
|
15822 |
0.13 |
| chr6_164518955_164519106 | 0.16 |
ENSG00000266128 |
. |
257437 |
0.02 |
| chr10_22764575_22765032 | 0.16 |
RP11-301N24.3 |
|
114702 |
0.06 |
| chr10_88852872_88853023 | 0.16 |
GLUD1 |
glutamate dehydrogenase 1 |
880 |
0.36 |
| chr12_25017223_25017374 | 0.16 |
RP11-625L16.3 |
|
33943 |
0.15 |
| chr11_2934551_2934724 | 0.16 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
9667 |
0.12 |
| chr4_78980714_78980865 | 0.16 |
FRAS1 |
Fraser syndrome 1 |
2065 |
0.46 |
| chr12_124871142_124871293 | 0.16 |
NCOR2 |
nuclear receptor corepressor 2 |
2153 |
0.43 |
| chr8_77493485_77493672 | 0.16 |
RP11-115I9.1 |
|
1998 |
0.42 |
| chr16_4579167_4579381 | 0.16 |
CDIP1 |
cell death-inducing p53 target 1 |
6748 |
0.13 |
| chrX_64254927_64255105 | 0.16 |
ZC4H2 |
zinc finger, C4H2 domain containing |
423 |
0.92 |
| chr7_76980949_76981220 | 0.16 |
GSAP |
gamma-secretase activating protein |
21400 |
0.22 |
| chr16_87379117_87379268 | 0.16 |
RP11-178L8.4 |
|
11313 |
0.1 |
| chr4_151000496_151000693 | 0.16 |
DCLK2 |
doublecortin-like kinase 2 |
414 |
0.91 |
| chr3_59503009_59503160 | 0.16 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
454499 |
0.01 |
| chr12_3358339_3358521 | 0.16 |
TSPAN9 |
tetraspanin 9 |
48089 |
0.16 |
| chr5_131543629_131543780 | 0.16 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
14080 |
0.16 |
| chr17_38600560_38600819 | 0.16 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
976 |
0.46 |
| chr8_144960232_144960383 | 0.16 |
EPPK1 |
epiplakin 1 |
7675 |
0.11 |
| chr2_23608807_23609156 | 0.16 |
KLHL29 |
kelch-like family member 29 |
893 |
0.74 |
| chr16_31488502_31488862 | 0.16 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
3935 |
0.1 |
| chr1_86968635_86969118 | 0.16 |
CLCA1 |
chloride channel accessory 1 |
34350 |
0.16 |
| chr17_42154780_42155022 | 0.16 |
G6PC3 |
glucose 6 phosphatase, catalytic, 3 |
2799 |
0.14 |
| chr4_113174579_113174763 | 0.15 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
11537 |
0.15 |
| chr11_61515322_61515473 | 0.15 |
MYRF |
myelin regulatory factor |
4724 |
0.15 |
| chr19_1907709_1907860 | 0.15 |
ADAT3 |
adenosine deaminase, tRNA-specific 3 |
234 |
0.68 |
| chrX_9801102_9801516 | 0.15 |
ENSG00000206844 |
. |
41432 |
0.15 |
| chr12_66232016_66232167 | 0.15 |
HMGA2 |
high mobility group AT-hook 2 |
13188 |
0.19 |
| chr16_88827167_88827318 | 0.15 |
RP5-1142A6.8 |
|
18013 |
0.07 |
| chr19_54386339_54386490 | 0.15 |
PRKCG |
protein kinase C, gamma |
499 |
0.56 |
| chr2_99552364_99552545 | 0.15 |
KIAA1211L |
KIAA1211-like |
212 |
0.96 |
| chr8_123874992_123875143 | 0.15 |
ZHX2 |
zinc fingers and homeoboxes 2 |
557 |
0.78 |
| chr11_45672012_45672287 | 0.15 |
RP11-495O11.1 |
|
930 |
0.62 |
| chr19_9879532_9879683 | 0.15 |
ZNF846 |
zinc finger protein 846 |
197 |
0.9 |
| chr12_194668_194819 | 0.15 |
IQSEC3 |
IQ motif and Sec7 domain 3 |
8201 |
0.17 |
| chr16_765719_765951 | 0.15 |
METRN |
meteorin, glial cell differentiation regulator |
194 |
0.8 |
| chr2_127826599_127826750 | 0.15 |
BIN1 |
bridging integrator 1 |
37903 |
0.18 |
| chr8_97825590_97825741 | 0.15 |
CPQ |
carboxypeptidase Q |
52197 |
0.18 |
| chr1_85527575_85527769 | 0.15 |
WDR63 |
WD repeat domain 63 |
323 |
0.89 |
| chr1_42046509_42046707 | 0.15 |
RP11-486B10.3 |
|
45494 |
0.16 |
| chr2_45162375_45162583 | 0.15 |
SIX3-AS1 |
SIX3 antisense RNA 1 |
6154 |
0.24 |
| chr19_17641532_17641686 | 0.15 |
FAM129C |
family with sequence similarity 129, member C |
3496 |
0.13 |
| chr2_105090262_105090413 | 0.15 |
ENSG00000199727 |
. |
58894 |
0.15 |
| chr7_151145465_151145709 | 0.15 |
CRYGN |
crystallin, gamma N |
8457 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.1 | 0.2 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.1 | 0.2 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.3 | GO:0045217 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.2 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:1903077 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 0.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.0 | GO:0032462 | regulation of protein homooligomerization(GO:0032462) |
| 0.0 | 0.1 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway(GO:0033145) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.2 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.0 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.0 | GO:0032347 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.0 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.0 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0031622 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.0 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0044254 | multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.2 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.0 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.0 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.0 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0042597 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.3 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.0 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.0 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |