Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
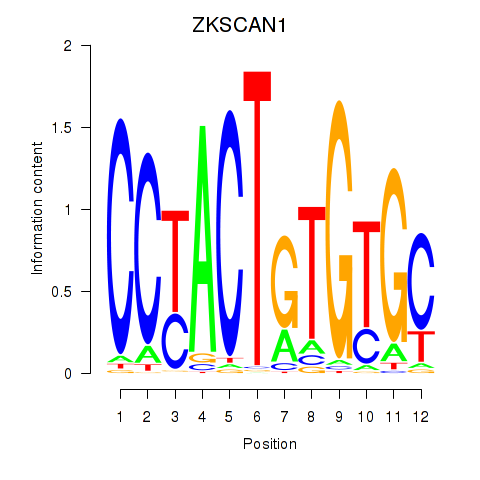
Results for ZKSCAN1
Z-value: 1.47
Transcription factors associated with ZKSCAN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN1
|
ENSG00000106261.12 | zinc finger with KRAB and SCAN domains 1 |
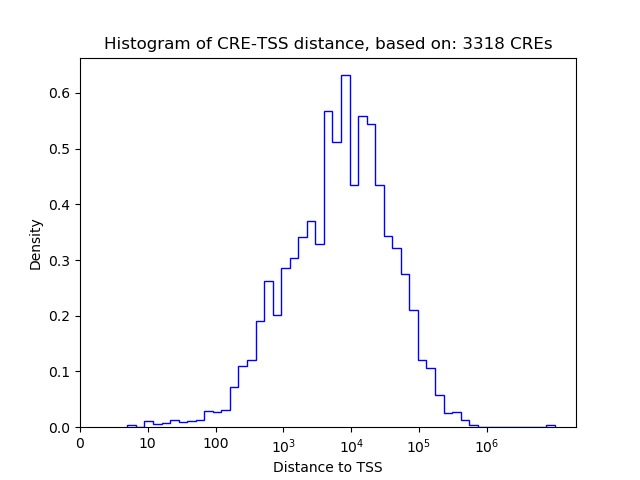
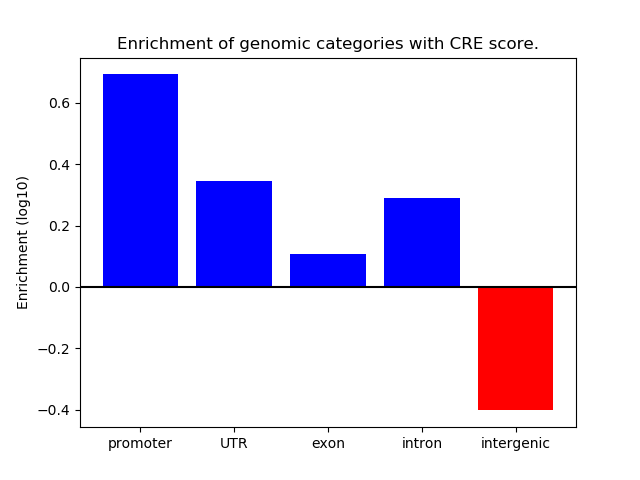
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_99595503_99596097 | ZKSCAN1 | 17404 | 0.083160 | 0.81 | 8.8e-03 | Click! |
| chr7_99614173_99614425 | ZKSCAN1 | 146 | 0.911082 | 0.67 | 4.7e-02 | Click! |
| chr7_99620138_99620289 | ZKSCAN1 | 5768 | 0.098537 | 0.63 | 6.7e-02 | Click! |
| chr7_99616377_99616588 | ZKSCAN1 | 2037 | 0.169780 | 0.63 | 7.1e-02 | Click! |
| chr7_99614433_99614621 | ZKSCAN1 | 82 | 0.938666 | 0.61 | 8.3e-02 | Click! |
Activity of the ZKSCAN1 motif across conditions
Conditions sorted by the z-value of the ZKSCAN1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
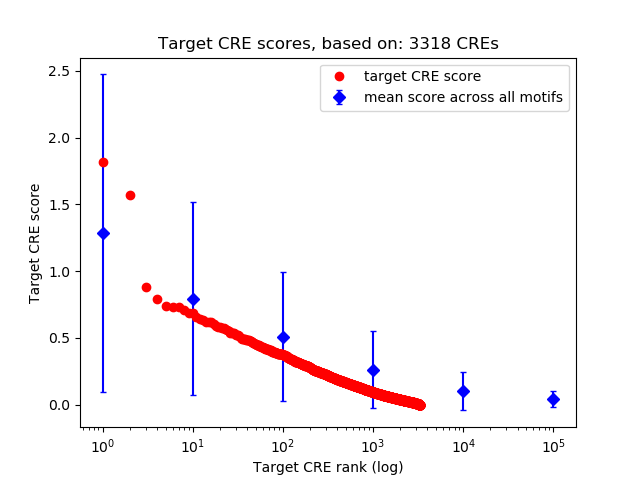
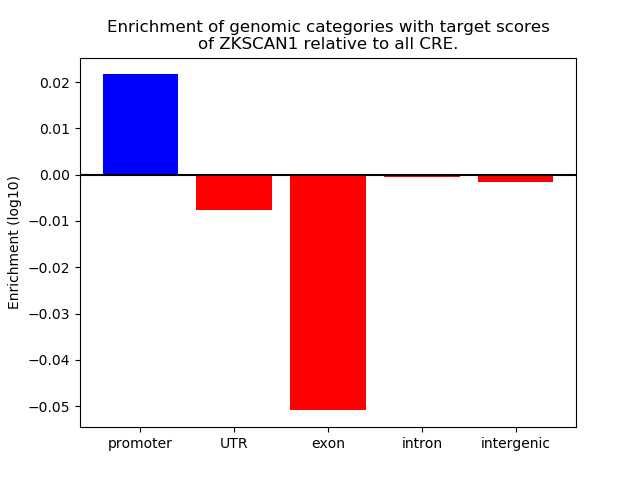
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_3876410_3876657 | 1.82 |
ATP2A3 |
ATPase, Ca++ transporting, ubiquitous |
8797 |
0.19 |
| chr18_74831342_74831493 | 1.57 |
MBP |
myelin basic protein |
8251 |
0.3 |
| chr2_437438_437589 | 0.88 |
FAM150B |
family with sequence similarity 150, member B |
148662 |
0.04 |
| chr10_11217517_11217809 | 0.79 |
RP3-323N1.2 |
|
4324 |
0.25 |
| chr19_42704508_42704707 | 0.74 |
ENSG00000265122 |
. |
7377 |
0.09 |
| chr2_231525959_231526216 | 0.73 |
CAB39 |
calcium binding protein 39 |
51473 |
0.12 |
| chr17_75877100_75877282 | 0.73 |
FLJ45079 |
|
1468 |
0.48 |
| chr3_13030755_13030906 | 0.71 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
2294 |
0.4 |
| chr1_198904768_198904950 | 0.69 |
ENSG00000207759 |
. |
76577 |
0.11 |
| chr7_949490_949865 | 0.69 |
ADAP1 |
ArfGAP with dual PH domains 1 |
4963 |
0.16 |
| chr1_203291694_203291901 | 0.66 |
ENSG00000202300 |
. |
3570 |
0.22 |
| chr1_23852013_23852546 | 0.64 |
E2F2 |
E2F transcription factor 2 |
5433 |
0.19 |
| chr17_21182635_21182786 | 0.64 |
MAP2K3 |
mitogen-activated protein kinase kinase 3 |
5274 |
0.19 |
| chr1_27943977_27944142 | 0.62 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
6514 |
0.15 |
| chr6_42397272_42397459 | 0.62 |
ENSG00000221252 |
. |
17231 |
0.19 |
| chr15_77312765_77312916 | 0.62 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
2041 |
0.33 |
| chr7_139311304_139311455 | 0.60 |
CLEC2L |
C-type lectin domain family 2, member L |
102551 |
0.07 |
| chr16_85481767_85482285 | 0.59 |
ENSG00000264203 |
. |
6928 |
0.27 |
| chr8_142415767_142415918 | 0.58 |
CTD-3064M3.4 |
|
13168 |
0.12 |
| chr7_50245194_50245403 | 0.58 |
AC020743.2 |
|
62879 |
0.12 |
| chr5_55884080_55884259 | 0.57 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
17890 |
0.22 |
| chr17_72752966_72753168 | 0.57 |
SLC9A3R1 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
5075 |
0.11 |
| chr6_11353172_11353323 | 0.56 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
29285 |
0.23 |
| chr11_117840834_117841225 | 0.56 |
IL10RA |
interleukin 10 receptor, alpha |
16034 |
0.17 |
| chr19_15782887_15783038 | 0.55 |
CYP4F12 |
cytochrome P450, family 4, subfamily F, polypeptide 12 |
605 |
0.72 |
| chr17_76125845_76126093 | 0.54 |
TMC6 |
transmembrane channel-like 6 |
795 |
0.36 |
| chr15_70773811_70774097 | 0.54 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
220666 |
0.02 |
| chr16_3146298_3146516 | 0.54 |
ZSCAN10 |
zinc finger and SCAN domain containing 10 |
2822 |
0.09 |
| chr20_62199024_62199397 | 0.53 |
HELZ2 |
helicase with zinc finger 2, transcriptional coactivator |
217 |
0.87 |
| chr14_91814952_91815599 | 0.52 |
ENSG00000265856 |
. |
15218 |
0.22 |
| chr14_65546707_65546994 | 0.52 |
RP11-840I19.3 |
|
1902 |
0.28 |
| chr2_198077243_198077408 | 0.52 |
ANKRD44 |
ankyrin repeat domain 44 |
14563 |
0.2 |
| chrX_128956937_128957088 | 0.51 |
ENSG00000266440 |
. |
1214 |
0.47 |
| chr7_97879296_97879465 | 0.50 |
TECPR1 |
tectonin beta-propeller repeat containing 1 |
1859 |
0.33 |
| chr15_44968349_44968500 | 0.49 |
PATL2 |
protein associated with topoisomerase II homolog 2 (yeast) |
324 |
0.85 |
| chr7_5701558_5701816 | 0.49 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
18405 |
0.18 |
| chr6_42902068_42902239 | 0.49 |
CNPY3 |
canopy FGF signaling regulator 3 |
5171 |
0.11 |
| chr10_6453263_6453585 | 0.49 |
DKFZP667F0711 |
|
61146 |
0.14 |
| chr1_198611512_198611708 | 0.49 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
3318 |
0.31 |
| chr3_12716668_12716819 | 0.48 |
RAF1 |
v-raf-1 murine leukemia viral oncogene homolog 1 |
11018 |
0.19 |
| chr3_3200095_3200246 | 0.48 |
RP11-97C16.1 |
|
5544 |
0.18 |
| chr1_203288364_203288515 | 0.48 |
ENSG00000202300 |
. |
212 |
0.93 |
| chr19_16476504_16477086 | 0.48 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
4031 |
0.18 |
| chr6_150048925_150049356 | 0.47 |
LATS1 |
large tumor suppressor kinase 1 |
9748 |
0.14 |
| chr1_12229589_12229973 | 0.47 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
2721 |
0.23 |
| chr14_98638353_98638504 | 0.47 |
ENSG00000222066 |
. |
159659 |
0.04 |
| chr11_65408846_65409255 | 0.46 |
SIPA1 |
signal-induced proliferation-associated 1 |
751 |
0.41 |
| chr1_12239932_12240089 | 0.46 |
ENSG00000263676 |
. |
11760 |
0.15 |
| chr5_169525252_169525403 | 0.46 |
FOXI1 |
forkhead box I1 |
7574 |
0.27 |
| chr20_48889044_48889195 | 0.46 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
81743 |
0.08 |
| chr4_40277829_40277980 | 0.45 |
CHRNA9 |
cholinergic receptor, nicotinic, alpha 9 (neuronal) |
59442 |
0.11 |
| chr19_15750443_15750594 | 0.45 |
CYP4F3 |
cytochrome P450, family 4, subfamily F, polypeptide 3 |
1189 |
0.47 |
| chr1_12211346_12211755 | 0.44 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
15510 |
0.15 |
| chr16_48654664_48654959 | 0.44 |
N4BP1 |
NEDD4 binding protein 1 |
10691 |
0.18 |
| chr1_175180542_175180693 | 0.44 |
KIAA0040 |
KIAA0040 |
18538 |
0.24 |
| chr8_38613649_38613800 | 0.44 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
1033 |
0.53 |
| chr10_126344624_126344775 | 0.43 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
47495 |
0.14 |
| chr19_2489875_2490034 | 0.43 |
ENSG00000252962 |
. |
13185 |
0.14 |
| chrX_19886161_19886348 | 0.43 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
19323 |
0.24 |
| chr9_136653979_136654130 | 0.43 |
SARDH |
sarcosine dehydrogenase |
48977 |
0.15 |
| chr10_14598436_14598650 | 0.43 |
FAM107B |
family with sequence similarity 107, member B |
364 |
0.91 |
| chr10_51499951_51500158 | 0.43 |
AGAP7 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 7 |
13727 |
0.2 |
| chr19_1086467_1086654 | 0.42 |
POLR2E |
polymerase (RNA) II (DNA directed) polypeptide E, 25kDa |
8793 |
0.09 |
| chr10_90586675_90586853 | 0.42 |
LIPM |
lipase, family member M |
24059 |
0.15 |
| chr1_208046544_208046843 | 0.42 |
CD34 |
CD34 molecule |
17144 |
0.25 |
| chr3_171080293_171080444 | 0.42 |
ENSG00000222506 |
. |
76947 |
0.11 |
| chr1_202127780_202127949 | 0.42 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
1255 |
0.37 |
| chr15_31563928_31564079 | 0.42 |
KLF13 |
Kruppel-like factor 13 |
55055 |
0.15 |
| chr12_50902770_50903124 | 0.41 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
4057 |
0.27 |
| chr5_176796790_176797014 | 0.41 |
RGS14 |
regulator of G-protein signaling 14 |
2958 |
0.13 |
| chr6_41240534_41240685 | 0.41 |
TREM1 |
triggering receptor expressed on myeloid cells 1 |
13794 |
0.13 |
| chr17_9966141_9966338 | 0.41 |
GAS7 |
growth arrest-specific 7 |
1543 |
0.42 |
| chr19_49121679_49121830 | 0.41 |
RPL18 |
ribosomal protein L18 |
613 |
0.39 |
| chr19_10831404_10831582 | 0.41 |
DNM2 |
dynamin 2 |
2256 |
0.18 |
| chr2_101260265_101260620 | 0.40 |
ENSG00000265839 |
. |
43114 |
0.15 |
| chr19_2274369_2274549 | 0.40 |
OAZ1 |
ornithine decarboxylase antizyme 1 |
4169 |
0.09 |
| chr17_18947608_18947759 | 0.40 |
GRAP |
GRB2-related adaptor protein |
1885 |
0.21 |
| chr7_5370188_5370339 | 0.40 |
TNRC18 |
trinucleotide repeat containing 18 |
2246 |
0.29 |
| chr13_99911046_99911220 | 0.39 |
GPR18 |
G protein-coupled receptor 18 |
451 |
0.83 |
| chr6_42015063_42015705 | 0.39 |
CCND3 |
cyclin D3 |
1040 |
0.46 |
| chr5_169706251_169706453 | 0.39 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
12021 |
0.22 |
| chr2_233083519_233083826 | 0.39 |
ENSG00000207626 |
. |
46309 |
0.14 |
| chr10_70856277_70856489 | 0.39 |
SRGN |
serglycin |
8509 |
0.19 |
| chr1_221951817_221951998 | 0.39 |
DUSP10 |
dual specificity phosphatase 10 |
36389 |
0.21 |
| chr5_177240356_177240599 | 0.39 |
FAM153A |
family with sequence similarity 153, member A |
30078 |
0.13 |
| chr10_90593085_90593348 | 0.38 |
ANKRD22 |
ankyrin repeat domain 22 |
18359 |
0.16 |
| chr22_27074499_27074686 | 0.38 |
CRYBA4 |
crystallin, beta A4 |
56664 |
0.12 |
| chr11_76091036_76091263 | 0.38 |
PRKRIR |
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) |
837 |
0.42 |
| chr9_137137039_137137190 | 0.38 |
RXRA |
retinoid X receptor, alpha |
81312 |
0.09 |
| chr11_67212387_67212841 | 0.38 |
CORO1B |
coronin, actin binding protein, 1B |
1351 |
0.18 |
| chr10_74081918_74082189 | 0.38 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
22666 |
0.16 |
| chr10_97597159_97597443 | 0.38 |
RP11-429G19.3 |
|
4036 |
0.26 |
| chr1_23925674_23925825 | 0.38 |
MDS2 |
myelodysplastic syndrome 2 translocation associated |
28075 |
0.14 |
| chr13_96335043_96335194 | 0.38 |
DNAJC3 |
DnaJ (Hsp40) homolog, subfamily C, member 3 |
5706 |
0.23 |
| chr20_31174503_31174689 | 0.38 |
RP11-410N8.4 |
|
685 |
0.59 |
| chr10_11740535_11740686 | 0.38 |
ECHDC3 |
enoyl CoA hydratase domain containing 3 |
43755 |
0.16 |
| chr12_57913214_57913504 | 0.38 |
ENSG00000208028 |
. |
317 |
0.62 |
| chr5_159913380_159913531 | 0.38 |
ENSG00000253522 |
. |
1096 |
0.49 |
| chr12_121272474_121272747 | 0.37 |
SPPL3 |
signal peptide peptidase like 3 |
850 |
0.61 |
| chr19_11517184_11517450 | 0.37 |
ENSG00000202357 |
. |
2131 |
0.14 |
| chr5_67562733_67562884 | 0.37 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
13260 |
0.27 |
| chr2_28880167_28880318 | 0.37 |
AC074011.2 |
|
7164 |
0.19 |
| chr2_114018866_114019227 | 0.37 |
ENSG00000189223 |
. |
6080 |
0.17 |
| chr9_134155011_134155162 | 0.37 |
FAM78A |
family with sequence similarity 78, member A |
3152 |
0.23 |
| chr16_3138152_3138303 | 0.37 |
ENSG00000200204 |
. |
2083 |
0.11 |
| chr5_134240040_134240274 | 0.37 |
PCBD2 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
439 |
0.8 |
| chr5_175457280_175457431 | 0.37 |
THOC3 |
THO complex 3 |
4328 |
0.17 |
| chr15_60863549_60863700 | 0.36 |
RORA |
RAR-related orphan receptor A |
21116 |
0.19 |
| chr11_48062772_48062967 | 0.36 |
AC103828.1 |
|
25462 |
0.18 |
| chr10_494842_494993 | 0.36 |
RP11-490E15.2 |
|
11416 |
0.2 |
| chr19_42376134_42376473 | 0.36 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
4887 |
0.12 |
| chr22_40450666_40450817 | 0.36 |
TNRC6B |
trinucleotide repeat containing 6B |
9263 |
0.19 |
| chr3_32179971_32180291 | 0.35 |
GPD1L |
glycerol-3-phosphate dehydrogenase 1-like |
31940 |
0.15 |
| chr3_71768573_71768891 | 0.35 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
5794 |
0.24 |
| chr1_54946013_54946309 | 0.35 |
ENSG00000265404 |
. |
24180 |
0.16 |
| chr17_71981136_71981299 | 0.35 |
RPL38 |
ribosomal protein L38 |
218504 |
0.02 |
| chr17_47471640_47471845 | 0.35 |
RP11-1079K10.4 |
|
9701 |
0.11 |
| chr10_80907133_80907284 | 0.35 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
78416 |
0.1 |
| chr19_3178230_3178381 | 0.35 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
431 |
0.74 |
| chr12_110290929_110291080 | 0.35 |
ENSG00000241413 |
. |
13310 |
0.13 |
| chr1_27940236_27940387 | 0.35 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
10168 |
0.14 |
| chr16_30192495_30192646 | 0.34 |
CORO1A |
coronin, actin binding protein, 1A |
1578 |
0.18 |
| chr11_46367187_46367549 | 0.34 |
DGKZ |
diacylglycerol kinase, zeta |
381 |
0.82 |
| chr11_75111382_75111533 | 0.34 |
RPS3 |
ribosomal protein S3 |
9 |
0.5 |
| chr22_40309826_40310035 | 0.34 |
GRAP2 |
GRB2-related adaptor protein 2 |
12665 |
0.17 |
| chr2_12318593_12318886 | 0.34 |
ENSG00000264089 |
. |
20517 |
0.27 |
| chr17_80821237_80821457 | 0.34 |
TBCD |
tubulin folding cofactor D |
3077 |
0.23 |
| chr15_31633419_31633718 | 0.34 |
KLF13 |
Kruppel-like factor 13 |
1432 |
0.57 |
| chr18_9153404_9153628 | 0.34 |
ANKRD12 |
ankyrin repeat domain 12 |
15955 |
0.15 |
| chr22_40527439_40527729 | 0.33 |
TNRC6B |
trinucleotide repeat containing 6B |
46345 |
0.15 |
| chr9_131998104_131998424 | 0.33 |
ENSG00000220992 |
. |
5268 |
0.19 |
| chr12_107766943_107767139 | 0.33 |
ENSG00000200897 |
. |
1483 |
0.51 |
| chr11_61197700_61198009 | 0.33 |
SDHAF2 |
succinate dehydrogenase complex assembly factor 2 |
238 |
0.44 |
| chr19_10746196_10746374 | 0.33 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
2035 |
0.19 |
| chr1_25970325_25970699 | 0.32 |
RP1-187B23.1 |
|
15090 |
0.18 |
| chr20_57733456_57733718 | 0.32 |
ZNF831 |
zinc finger protein 831 |
32488 |
0.19 |
| chr11_73091795_73092090 | 0.32 |
RELT |
RELT tumor necrosis factor receptor |
4229 |
0.17 |
| chr17_33221100_33221251 | 0.32 |
CCT6B |
chaperonin containing TCP1, subunit 6B (zeta 2) |
67246 |
0.09 |
| chr17_73649033_73649246 | 0.32 |
SMIM6 |
small integral membrane protein 6 |
6622 |
0.1 |
| chr11_6423240_6423565 | 0.32 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
2492 |
0.21 |
| chr2_46926793_46927032 | 0.32 |
SOCS5 |
suppressor of cytokine signaling 5 |
586 |
0.78 |
| chr17_65434813_65435235 | 0.32 |
ENSG00000244610 |
. |
18382 |
0.13 |
| chr4_6941984_6942245 | 0.32 |
ENSG00000265953 |
. |
12902 |
0.15 |
| chr22_27083769_27083920 | 0.32 |
CRYBA4 |
crystallin, beta A4 |
65916 |
0.11 |
| chr10_114141969_114142120 | 0.32 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
6087 |
0.2 |
| chr1_54814987_54815138 | 0.32 |
SSBP3 |
single stranded DNA binding protein 3 |
56115 |
0.11 |
| chr9_137281945_137282096 | 0.32 |
ENSG00000263897 |
. |
10763 |
0.23 |
| chr1_225608177_225608328 | 0.32 |
LBR |
lamin B receptor |
7347 |
0.19 |
| chr3_46278155_46278310 | 0.31 |
CCR3 |
chemokine (C-C motif) receptor 3 |
5640 |
0.23 |
| chr19_54722925_54723182 | 0.31 |
LILRB3 |
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 |
3797 |
0.09 |
| chr17_56403632_56403944 | 0.31 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
977 |
0.36 |
| chr1_53703818_53703969 | 0.31 |
MAGOH |
mago-nashi homolog, proliferation-associated (Drosophila) |
278 |
0.56 |
| chr16_75094649_75094937 | 0.31 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
43889 |
0.12 |
| chr19_18285022_18285173 | 0.31 |
IFI30 |
interferon, gamma-inducible protein 30 |
620 |
0.57 |
| chr16_4364379_4364530 | 0.31 |
GLIS2 |
GLIS family zinc finger 2 |
308 |
0.85 |
| chr1_111203801_111203979 | 0.31 |
ENSG00000221538 |
. |
8237 |
0.18 |
| chr3_15369856_15370007 | 0.31 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
4136 |
0.17 |
| chr16_17545376_17545755 | 0.31 |
XYLT1 |
xylosyltransferase I |
19173 |
0.3 |
| chr14_74241491_74241738 | 0.30 |
ELMSAN1 |
ELM2 and Myb/SANT-like domain containing 1 |
12347 |
0.11 |
| chr1_247332601_247332804 | 0.30 |
ZNF124 |
zinc finger protein 124 |
2567 |
0.22 |
| chr12_3863154_3863452 | 0.30 |
EFCAB4B |
EF-hand calcium binding domain 4B |
937 |
0.65 |
| chr18_13219395_13219546 | 0.30 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
684 |
0.66 |
| chr14_103113093_103113249 | 0.30 |
ENSG00000202459 |
. |
48554 |
0.11 |
| chr2_8201184_8201335 | 0.30 |
ENSG00000221255 |
. |
484287 |
0.01 |
| chr5_173202423_173202604 | 0.30 |
ENSG00000263401 |
. |
45432 |
0.18 |
| chr20_31129022_31129283 | 0.30 |
C20orf112 |
chromosome 20 open reading frame 112 |
4952 |
0.19 |
| chr11_60789970_60790185 | 0.30 |
CD6 |
CD6 molecule |
14071 |
0.14 |
| chr22_40336508_40336741 | 0.30 |
GRAP2 |
GRB2-related adaptor protein 2 |
6197 |
0.18 |
| chr14_92912996_92913173 | 0.30 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
7376 |
0.28 |
| chr9_135897951_135898139 | 0.30 |
GTF3C5 |
general transcription factor IIIC, polypeptide 5, 63kDa |
8031 |
0.14 |
| chr14_60693586_60693737 | 0.30 |
CTD-2184C24.2 |
|
18393 |
0.19 |
| chr19_42956472_42956792 | 0.30 |
LIPE-AS1 |
LIPE antisense RNA 1 |
8368 |
0.12 |
| chr2_213879689_213879840 | 0.30 |
AC079610.1 |
|
6505 |
0.31 |
| chr13_25202028_25202179 | 0.30 |
ENSG00000211508 |
. |
18886 |
0.18 |
| chr14_54977048_54977291 | 0.30 |
CGRRF1 |
cell growth regulator with ring finger domain 1 |
545 |
0.79 |
| chr18_21582787_21583054 | 0.30 |
TTC39C |
tetratricopeptide repeat domain 39C |
10183 |
0.18 |
| chr3_50653123_50653503 | 0.29 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
1249 |
0.38 |
| chr10_11131193_11131344 | 0.29 |
CELF2-AS2 |
CELF2 antisense RNA 2 |
8855 |
0.23 |
| chr21_46047532_46047683 | 0.29 |
KRTAP10-9 |
keratin associated protein 10-9 |
567 |
0.52 |
| chr20_31056876_31057055 | 0.29 |
C20orf112 |
chromosome 20 open reading frame 112 |
14309 |
0.17 |
| chr11_94997219_94997453 | 0.29 |
RP11-712B9.2 |
|
31601 |
0.19 |
| chr12_93492296_93492447 | 0.29 |
ENSG00000221617 |
. |
1344 |
0.49 |
| chr3_13021430_13021581 | 0.29 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
7031 |
0.28 |
| chr7_155785095_155785246 | 0.29 |
AC021218.2 |
Uncharacterized protein |
29844 |
0.23 |
| chr19_9946433_9946665 | 0.29 |
PIN1 |
peptidylprolyl cis/trans isomerase, NIMA-interacting 1 |
532 |
0.58 |
| chr19_8400659_8400845 | 0.29 |
KANK3 |
KN motif and ankyrin repeat domains 3 |
7394 |
0.1 |
| chr1_226835966_226836117 | 0.29 |
ITPKB-IT1 |
ITPKB intronic transcript 1 (non-protein coding) |
11330 |
0.21 |
| chr11_48018118_48018305 | 0.29 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
15932 |
0.19 |
| chr12_89638036_89638195 | 0.29 |
ENSG00000238302 |
. |
37947 |
0.21 |
| chr6_126230387_126230538 | 0.29 |
NCOA7 |
nuclear receptor coactivator 7 |
9407 |
0.19 |
| chr11_65552908_65553161 | 0.29 |
OVOL1 |
ovo-like zinc finger 1 |
1459 |
0.22 |
| chr12_105044364_105044605 | 0.29 |
ENSG00000264295 |
. |
59073 |
0.13 |
| chr19_7929180_7929331 | 0.29 |
EVI5L |
ecotropic viral integration site 5-like |
3784 |
0.1 |
| chrX_12972828_12973236 | 0.28 |
TMSB4X |
thymosin beta 4, X-linked |
20195 |
0.19 |
| chr8_131020536_131020825 | 0.28 |
ENSG00000264653 |
. |
19 |
0.97 |
| chr1_1827398_1827549 | 0.28 |
RP1-140A9.1 |
|
4563 |
0.14 |
| chr14_70165157_70165557 | 0.28 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
28260 |
0.2 |
| chr15_77291917_77292573 | 0.28 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
4330 |
0.22 |
| chr16_17475953_17476212 | 0.28 |
XYLT1 |
xylosyltransferase I |
88656 |
0.11 |
| chr20_1946593_1946814 | 0.28 |
RP4-684O24.5 |
|
18811 |
0.19 |
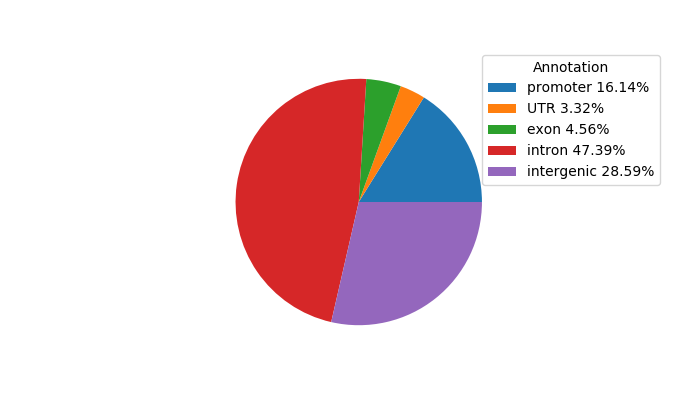
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.1 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.1 | 0.2 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.1 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0072338 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:1901985 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.2 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.0 | GO:0070423 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.2 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:2001021 | negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 | 0.1 | GO:0035844 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:1903313 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.4 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.0 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.6 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.0 | 0.1 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0048617 | embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.0 | 0.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0045683 | negative regulation of epidermal cell differentiation(GO:0045605) negative regulation of epidermis development(GO:0045683) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.3 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.2 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0051788 | response to misfolded protein(GO:0051788) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.0 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) membrane raft localization(GO:0051665) |
| 0.0 | 0.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0043301 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0042346 | positive regulation of NF-kappaB import into nucleus(GO:0042346) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.0 | GO:0051446 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0042094 | interleukin-2 biosynthetic process(GO:0042094) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.3 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 0.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.0 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) |
| 0.0 | 0.1 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.3 | GO:0051318 | G1 phase(GO:0051318) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.0 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.0 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.0 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.0 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.0 | REACTOME SIGNAL AMPLIFICATION | Genes involved in Signal amplification |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |