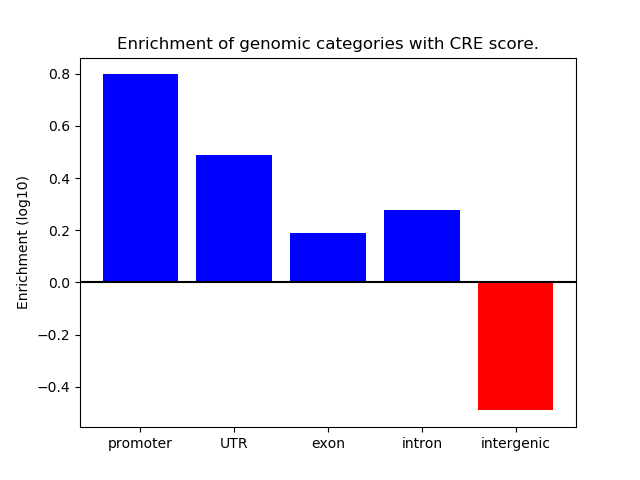
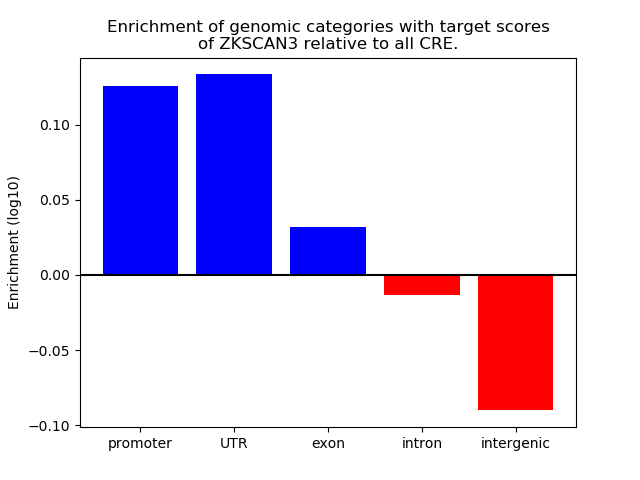
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
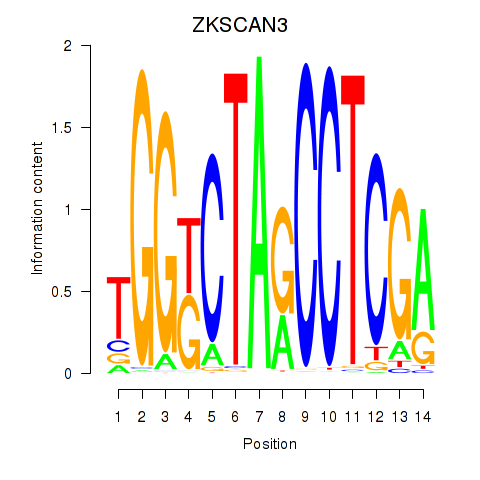
Results for ZKSCAN3
Z-value: 1.44
Transcription factors associated with ZKSCAN3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN3
|
ENSG00000189298.9 | zinc finger with KRAB and SCAN domains 3 |
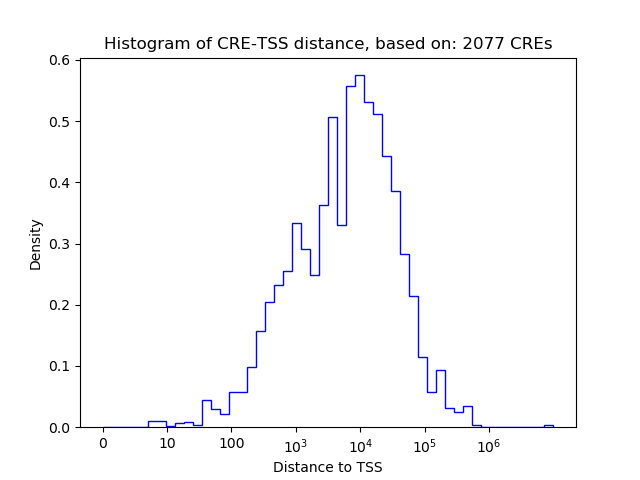
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_28318404_28318713 | ZKSCAN3 | 831 | 0.531891 | 0.66 | 5.5e-02 | Click! |
| chr6_28318919_28319190 | ZKSCAN3 | 1327 | 0.348212 | 0.54 | 1.3e-01 | Click! |
| chr6_28318026_28318177 | ZKSCAN3 | 374 | 0.819170 | 0.44 | 2.3e-01 | Click! |
| chr6_28317244_28317413 | ZKSCAN3 | 363 | 0.828791 | 0.27 | 4.8e-01 | Click! |
| chr6_28317523_28317674 | ZKSCAN3 | 93 | 0.958716 | 0.05 | 8.9e-01 | Click! |
Activity of the ZKSCAN3 motif across conditions
Conditions sorted by the z-value of the ZKSCAN3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
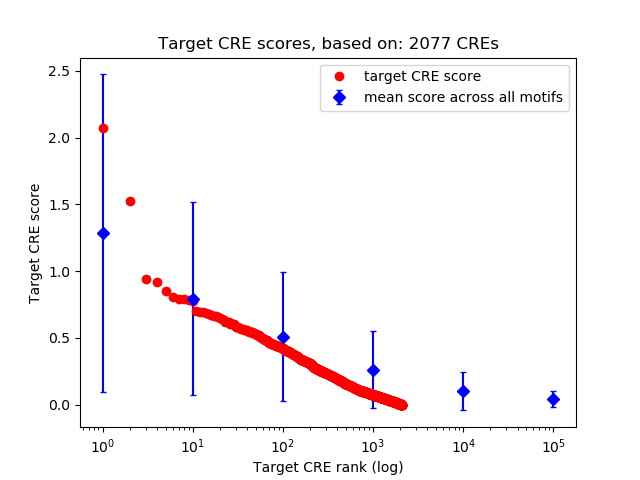
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_14610353_14610666 | 2.07 |
FAM107B |
family with sequence similarity 107, member B |
3520 |
0.31 |
| chr9_100866127_100866462 | 1.53 |
TRIM14 |
tripartite motif containing 14 |
11451 |
0.18 |
| chr11_64013353_64013610 | 0.94 |
RP11-783K16.5 |
|
45 |
0.65 |
| chr1_211509622_211509899 | 0.92 |
TRAF5 |
TNF receptor-associated factor 5 |
9581 |
0.25 |
| chr1_26100660_26100867 | 0.85 |
SEPN1 |
selenoprotein N, 1 |
25904 |
0.11 |
| chr14_50563249_50563678 | 0.81 |
C14orf183 |
chromosome 14 open reading frame 183 |
4102 |
0.18 |
| chr8_37748769_37748986 | 0.79 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
8095 |
0.14 |
| chr4_100766407_100766703 | 0.79 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
28552 |
0.18 |
| chr14_93159663_93159814 | 0.78 |
LGMN |
legumain |
24066 |
0.21 |
| chr1_92940802_92940965 | 0.78 |
GFI1 |
growth factor independent 1 transcription repressor |
8628 |
0.26 |
| chr2_227702908_227703080 | 0.70 |
RHBDD1 |
rhomboid domain containing 1 |
730 |
0.71 |
| chr1_25361714_25361887 | 0.70 |
ENSG00000264371 |
. |
11806 |
0.23 |
| chr16_57056402_57056553 | 0.69 |
NLRC5 |
NLR family, CARD domain containing 5 |
1852 |
0.27 |
| chr2_97167178_97167382 | 0.69 |
NEURL3 |
neuralized E3 ubiquitin protein ligase 3 |
619 |
0.73 |
| chr20_4789979_4790248 | 0.68 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
5656 |
0.22 |
| chr20_56193901_56194158 | 0.67 |
ZBP1 |
Z-DNA binding protein 1 |
1421 |
0.48 |
| chr1_211526157_211526486 | 0.67 |
TRAF5 |
TNF receptor-associated factor 5 |
6615 |
0.27 |
| chr11_73680269_73680439 | 0.66 |
RP11-167N4.2 |
|
978 |
0.46 |
| chr15_60688686_60688869 | 0.65 |
ANXA2 |
annexin A2 |
760 |
0.75 |
| chr15_64720015_64720166 | 0.65 |
TRIP4 |
thyroid hormone receptor interactor 4 |
3776 |
0.14 |
| chr9_135363597_135363846 | 0.64 |
ENSG00000252521 |
. |
66693 |
0.1 |
| chr22_40327971_40328220 | 0.63 |
GRAP2 |
GRB2-related adaptor protein 2 |
5454 |
0.19 |
| chr17_71589495_71589717 | 0.62 |
RP11-277J6.2 |
|
47559 |
0.14 |
| chr11_73684050_73684451 | 0.62 |
RP11-167N4.2 |
|
2918 |
0.19 |
| chr12_14577482_14577735 | 0.62 |
ATF7IP |
activating transcription factor 7 interacting protein |
913 |
0.66 |
| chr10_15009390_15009541 | 0.61 |
MEIG1 |
meiosis/spermiogenesis associated 1 |
1027 |
0.53 |
| chr2_136891083_136891299 | 0.61 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
15456 |
0.27 |
| chr19_682192_682343 | 0.61 |
FSTL3 |
follistatin-like 3 (secreted glycoprotein) |
4325 |
0.11 |
| chr14_99678734_99678885 | 0.61 |
AL109767.1 |
|
50476 |
0.13 |
| chr4_89517359_89517510 | 0.58 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
2905 |
0.24 |
| chr11_73095605_73095891 | 0.58 |
RELT |
RELT tumor necrosis factor receptor |
8035 |
0.15 |
| chr5_118607191_118607349 | 0.58 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
2821 |
0.26 |
| chr3_9821453_9821979 | 0.58 |
CAMK1 |
calcium/calmodulin-dependent protein kinase I |
10040 |
0.09 |
| chr1_101749629_101749820 | 0.57 |
RP4-575N6.5 |
|
41010 |
0.13 |
| chr1_67727636_67727815 | 0.57 |
ENSG00000252116 |
. |
5684 |
0.17 |
| chr7_150178587_150178758 | 0.56 |
GIMAP8 |
GTPase, IMAP family member 8 |
30954 |
0.13 |
| chr15_28382300_28382484 | 0.56 |
HERC2 |
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
37177 |
0.18 |
| chr3_185313043_185313423 | 0.56 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
9150 |
0.23 |
| chr1_26873022_26873700 | 0.56 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
1018 |
0.45 |
| chr9_137039448_137039599 | 0.56 |
ENSG00000221676 |
. |
9837 |
0.2 |
| chr6_112133106_112133257 | 0.55 |
FYN |
FYN oncogene related to SRC, FGR, YES |
8100 |
0.29 |
| chr13_30950886_30951061 | 0.55 |
KATNAL1 |
katanin p60 subunit A-like 1 |
69352 |
0.11 |
| chr3_15503071_15503367 | 0.55 |
EAF1-AS1 |
EAF1 antisense RNA 1 |
10110 |
0.12 |
| chr17_60872822_60872973 | 0.55 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
11104 |
0.24 |
| chr7_37382804_37383115 | 0.55 |
ELMO1 |
engulfment and cell motility 1 |
176 |
0.96 |
| chr9_139423606_139423897 | 0.54 |
ENSG00000263403 |
. |
9673 |
0.09 |
| chr19_7702843_7703102 | 0.53 |
STXBP2 |
syntaxin binding protein 2 |
959 |
0.26 |
| chr15_55514367_55514559 | 0.53 |
RSL24D1 |
ribosomal L24 domain containing 1 |
25198 |
0.18 |
| chr14_61831663_61831831 | 0.53 |
PRKCH |
protein kinase C, eta |
3955 |
0.27 |
| chr15_31134475_31134626 | 0.53 |
ENSG00000221379 |
. |
41041 |
0.12 |
| chr14_56582946_56583243 | 0.52 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
1438 |
0.49 |
| chr14_35303784_35304022 | 0.52 |
ENSG00000251726 |
. |
10663 |
0.17 |
| chr2_241562052_241562405 | 0.52 |
GPR35 |
G protein-coupled receptor 35 |
2434 |
0.21 |
| chr11_44631349_44631561 | 0.52 |
RP11-58K22.4 |
|
1804 |
0.34 |
| chr9_20378989_20379416 | 0.51 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
3265 |
0.26 |
| chr20_8116239_8116390 | 0.51 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
3012 |
0.36 |
| chr15_64260424_64260713 | 0.51 |
RP11-111E14.1 |
|
40095 |
0.15 |
| chr9_129159177_129159360 | 0.50 |
ENSG00000253079 |
. |
11421 |
0.2 |
| chr14_77234089_77234240 | 0.50 |
VASH1 |
vasohibin 1 |
5492 |
0.2 |
| chr20_55059757_55060025 | 0.49 |
ENSG00000238294 |
. |
9192 |
0.13 |
| chr13_41576048_41576351 | 0.49 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
17251 |
0.19 |
| chr19_6766080_6766650 | 0.49 |
SH2D3A |
SH2 domain containing 3A |
1081 |
0.38 |
| chr3_170942262_170942413 | 0.49 |
TNIK |
TRAF2 and NCK interacting kinase |
1163 |
0.63 |
| chrX_53740602_53740909 | 0.49 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
27082 |
0.23 |
| chr19_39728024_39728175 | 0.48 |
IFNL3 |
interferon, lambda 3 |
7547 |
0.12 |
| chr3_183259605_183259756 | 0.48 |
KLHL6-AS1 |
KLHL6 antisense RNA 1 |
6843 |
0.17 |
| chr12_57865222_57865373 | 0.48 |
ARHGAP9 |
Rho GTPase activating protein 9 |
3420 |
0.1 |
| chr14_34512264_34512415 | 0.48 |
EGLN3-AS1 |
EGLN3 antisense RNA 1 |
3036 |
0.35 |
| chr17_7779861_7780054 | 0.47 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
8167 |
0.09 |
| chr14_89977838_89978024 | 0.47 |
FOXN3 |
forkhead box N3 |
17402 |
0.19 |
| chr4_6916161_6916458 | 0.46 |
TBC1D14 |
TBC1 domain family, member 14 |
4334 |
0.22 |
| chr7_4748966_4749117 | 0.46 |
ENSG00000238781 |
. |
19930 |
0.18 |
| chr17_42326114_42326265 | 0.46 |
AC003102.1 |
|
1047 |
0.29 |
| chr2_39472617_39472990 | 0.46 |
CDKL4 |
cyclin-dependent kinase-like 4 |
16130 |
0.19 |
| chr2_118771117_118771268 | 0.46 |
CCDC93 |
coiled-coil domain containing 93 |
517 |
0.7 |
| chr8_818924_819075 | 0.46 |
ERICH1-AS1 |
ERICH1 antisense RNA 1 |
14199 |
0.29 |
| chr5_14587040_14587260 | 0.45 |
FAM105A |
family with sequence similarity 105, member A |
5266 |
0.28 |
| chr21_47335720_47335871 | 0.45 |
PRED62 |
Uncharacterized protein |
16682 |
0.18 |
| chr20_1677648_1677799 | 0.45 |
ENSG00000242348 |
. |
30717 |
0.14 |
| chr3_122512892_122513151 | 0.45 |
HSPBAP1 |
HSPB (heat shock 27kDa) associated protein 1 |
350 |
0.67 |
| chr17_17209430_17209581 | 0.45 |
NT5M |
5',3'-nucleotidase, mitochondrial |
2856 |
0.24 |
| chr3_52267134_52267345 | 0.45 |
TLR9 |
TLR9 |
2033 |
0.17 |
| chr11_35073040_35073191 | 0.45 |
PDHX |
pyruvate dehydrogenase complex, component X |
73784 |
0.08 |
| chr14_22886920_22887071 | 0.44 |
ENSG00000251002 |
. |
14724 |
0.12 |
| chr21_47971487_47971638 | 0.44 |
ENSG00000272283 |
. |
15853 |
0.17 |
| chr7_138777169_138777799 | 0.44 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
13471 |
0.2 |
| chr1_1148836_1149071 | 0.44 |
TNFRSF4 |
tumor necrosis factor receptor superfamily, member 4 |
559 |
0.53 |
| chr18_13313200_13313369 | 0.44 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
35182 |
0.14 |
| chr9_34122816_34123204 | 0.44 |
DCAF12 |
DDB1 and CUL4 associated factor 12 |
2715 |
0.22 |
| chr18_21601039_21601542 | 0.43 |
TTC39C |
tetratricopeptide repeat domain 39C |
4692 |
0.2 |
| chr1_65408943_65409111 | 0.43 |
JAK1 |
Janus kinase 1 |
23160 |
0.22 |
| chr1_160654931_160655551 | 0.43 |
RP11-404F10.2 |
|
11929 |
0.15 |
| chr21_34561335_34561486 | 0.43 |
C21orf54 |
chromosome 21 open reading frame 54 |
18869 |
0.14 |
| chr4_2837699_2838048 | 0.43 |
ADD1 |
adducin 1 (alpha) |
7711 |
0.2 |
| chr15_60883775_60883968 | 0.43 |
RORA |
RAR-related orphan receptor A |
869 |
0.66 |
| chr12_62998741_62998934 | 0.43 |
ENSG00000199179 |
. |
1371 |
0.28 |
| chr1_205884511_205884662 | 0.43 |
RP4-681L3.2 |
|
19670 |
0.17 |
| chr1_67426336_67426573 | 0.43 |
MIER1 |
mesoderm induction early response 1, transcriptional regulator |
30528 |
0.16 |
| chr21_36391287_36391723 | 0.43 |
RUNX1 |
runt-related transcription factor 1 |
29957 |
0.26 |
| chr5_56025506_56025889 | 0.42 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
85704 |
0.08 |
| chr19_19630373_19630524 | 0.42 |
NDUFA13 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
3409 |
0.12 |
| chr1_89667140_89667505 | 0.42 |
GBP4 |
guanylate binding protein 4 |
2707 |
0.26 |
| chr4_6918676_6918943 | 0.42 |
TBC1D14 |
TBC1 domain family, member 14 |
6834 |
0.19 |
| chr1_98380116_98380267 | 0.41 |
DPYD |
dihydropyrimidine dehydrogenase |
6362 |
0.32 |
| chr5_11234617_11234797 | 0.41 |
ENSG00000207312 |
. |
207395 |
0.03 |
| chr7_134852339_134852490 | 0.41 |
RP11-134L10.1 |
|
741 |
0.55 |
| chr16_25077360_25077511 | 0.41 |
LCMT1 |
leucine carboxyl methyltransferase 1 |
45615 |
0.14 |
| chr17_2236873_2237024 | 0.41 |
TSR1 |
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
2803 |
0.13 |
| chr22_21960363_21960532 | 0.41 |
YDJC |
YdjC homolog (bacterial) |
23878 |
0.08 |
| chr15_66065362_66065523 | 0.40 |
RP11-16E23.3 |
|
5706 |
0.18 |
| chr13_42895024_42895175 | 0.40 |
AKAP11 |
A kinase (PRKA) anchor protein 11 |
48810 |
0.18 |
| chr1_231171258_231171409 | 0.40 |
FAM89A |
family with sequence similarity 89, member A |
4659 |
0.18 |
| chr1_12242763_12243127 | 0.40 |
ENSG00000263676 |
. |
8825 |
0.16 |
| chr12_7063873_7064421 | 0.40 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
3609 |
0.07 |
| chr20_58515754_58515905 | 0.40 |
FAM217B |
family with sequence similarity 217, member B |
335 |
0.59 |
| chr13_99952051_99952321 | 0.40 |
GPR183 |
G protein-coupled receptor 183 |
7473 |
0.21 |
| chr1_206809391_206809612 | 0.40 |
DYRK3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
327 |
0.63 |
| chr17_58041513_58041664 | 0.40 |
RNFT1 |
ring finger protein, transmembrane 1 |
492 |
0.75 |
| chr9_97429931_97430112 | 0.40 |
FBP1 |
fructose-1,6-bisphosphatase 1 |
27490 |
0.19 |
| chr3_43236741_43236892 | 0.39 |
ENSG00000222331 |
. |
15547 |
0.22 |
| chr1_175166883_175167095 | 0.39 |
KIAA0040 |
KIAA0040 |
4910 |
0.3 |
| chr9_114426841_114427100 | 0.39 |
GNG10 |
guanine nucleotide binding protein (G protein), gamma 10 |
3355 |
0.21 |
| chr17_44218651_44218858 | 0.39 |
ENSG00000252698 |
. |
13992 |
0.17 |
| chr2_205834532_205835011 | 0.38 |
PARD3B |
par-3 family cell polarity regulator beta |
424048 |
0.01 |
| chr9_126763063_126763261 | 0.38 |
LHX2 |
LIM homeobox 2 |
787 |
0.6 |
| chr11_117198965_117199116 | 0.38 |
CEP164 |
centrosomal protein 164kDa |
221 |
0.57 |
| chrX_153231291_153231484 | 0.38 |
HCFC1-AS1 |
HCFC1 antisense RNA 1 |
3389 |
0.11 |
| chr15_42181166_42181317 | 0.38 |
RP11-23P13.6 |
|
3750 |
0.13 |
| chr15_60828599_60828948 | 0.38 |
CTD-2501E16.2 |
|
6601 |
0.21 |
| chr17_56361902_56362063 | 0.38 |
MPO |
myeloperoxidase |
3686 |
0.16 |
| chr15_30456809_30456986 | 0.38 |
ENSG00000221785 |
. |
19817 |
0.12 |
| chr11_32610462_32610653 | 0.38 |
EIF3M |
eukaryotic translation initiation factor 3, subunit M |
575 |
0.82 |
| chr11_1606465_1606821 | 0.37 |
KRTAP5-1 |
keratin associated protein 5-1 |
130 |
0.93 |
| chr1_47181224_47181375 | 0.37 |
EFCAB14 |
EF-hand calcium binding domain 14 |
3437 |
0.21 |
| chr7_96651846_96652002 | 0.37 |
DLX5 |
distal-less homeobox 5 |
2338 |
0.25 |
| chr1_35897155_35897306 | 0.37 |
ENSG00000202129 |
. |
3736 |
0.22 |
| chr8_53851916_53852182 | 0.37 |
NPBWR1 |
neuropeptides B/W receptor 1 |
1058 |
0.63 |
| chr5_171061383_171061534 | 0.37 |
ENSG00000264303 |
. |
136076 |
0.04 |
| chr6_35653753_35653924 | 0.37 |
FKBP5 |
FK506 binding protein 5 |
2854 |
0.19 |
| chr4_16124268_16124434 | 0.36 |
PROM1 |
prominin 1 |
38350 |
0.16 |
| chr3_59995792_59996170 | 0.36 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
38398 |
0.24 |
| chr15_32665699_32665938 | 0.36 |
ENSG00000221444 |
. |
19855 |
0.11 |
| chr19_4638499_4638663 | 0.36 |
TNFAIP8L1 |
tumor necrosis factor, alpha-induced protein 8-like 1 |
949 |
0.43 |
| chr6_24918083_24918272 | 0.36 |
FAM65B |
family with sequence similarity 65, member B |
6982 |
0.24 |
| chr3_9440127_9440389 | 0.36 |
SETD5-AS1 |
SETD5 antisense RNA 1 |
5 |
0.93 |
| chr3_9885266_9885439 | 0.36 |
RPUSD3 |
RNA pseudouridylate synthase domain containing 3 |
274 |
0.82 |
| chr20_35573807_35573970 | 0.36 |
SAMHD1 |
SAM domain and HD domain 1 |
6223 |
0.23 |
| chr20_30607376_30607540 | 0.35 |
CCM2L |
cerebral cavernous malformation 2-like |
437 |
0.74 |
| chr2_65295203_65295373 | 0.35 |
CEP68 |
centrosomal protein 68kDa |
11689 |
0.17 |
| chr15_30472324_30472499 | 0.35 |
ENSG00000221785 |
. |
35331 |
0.11 |
| chr1_90206501_90206652 | 0.35 |
ENSG00000239176 |
. |
27301 |
0.18 |
| chr10_32216291_32216562 | 0.35 |
ARHGAP12 |
Rho GTPase activating protein 12 |
1302 |
0.55 |
| chr7_142500079_142500230 | 0.35 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
19023 |
0.16 |
| chr2_106394710_106394861 | 0.34 |
NCK2 |
NCK adaptor protein 2 |
32597 |
0.22 |
| chr2_161945197_161945348 | 0.34 |
TANK |
TRAF family member-associated NFKB activator |
48147 |
0.16 |
| chr17_56394632_56394843 | 0.34 |
BZRAP1 |
benzodiazepine receptor (peripheral) associated protein 1 |
6636 |
0.12 |
| chr6_149794191_149794357 | 0.34 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
9995 |
0.17 |
| chr21_43948338_43948579 | 0.34 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
13824 |
0.13 |
| chr11_114030278_114030531 | 0.34 |
ENSG00000221112 |
. |
72753 |
0.1 |
| chr11_125461625_125461796 | 0.33 |
STT3A |
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
103 |
0.9 |
| chr18_8602838_8602989 | 0.33 |
RAB12 |
RAB12, member RAS oncogene family |
6530 |
0.22 |
| chr9_34981335_34981528 | 0.33 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
8207 |
0.15 |
| chr6_15399373_15399540 | 0.33 |
JARID2 |
jumonji, AT rich interactive domain 2 |
1633 |
0.48 |
| chr2_240673435_240673899 | 0.33 |
AC093802.1 |
Uncharacterized protein |
10887 |
0.29 |
| chr2_61243941_61244148 | 0.33 |
PUS10 |
pseudouridylate synthase 10 |
284 |
0.51 |
| chr19_21646642_21646886 | 0.33 |
ZNF429 |
zinc finger protein 429 |
41602 |
0.16 |
| chr1_27686480_27686799 | 0.33 |
MAP3K6 |
mitogen-activated protein kinase kinase kinase 6 |
3677 |
0.14 |
| chrX_43295883_43296068 | 0.33 |
MAOA |
monoamine oxidase A |
219492 |
0.02 |
| chr12_53024160_53024311 | 0.33 |
KRT73 |
keratin 73 |
11892 |
0.11 |
| chr2_103036026_103036177 | 0.33 |
IL18RAP |
interleukin 18 receptor accessory protein |
45 |
0.97 |
| chr1_26635107_26635385 | 0.33 |
UBXN11 |
UBX domain protein 11 |
1766 |
0.23 |
| chr16_75123794_75123945 | 0.33 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
14813 |
0.15 |
| chr4_78079689_78079921 | 0.33 |
CCNG2 |
cyclin G2 |
226 |
0.94 |
| chr12_12028552_12028703 | 0.33 |
ETV6 |
ets variant 6 |
10244 |
0.3 |
| chr3_5047856_5048016 | 0.33 |
BHLHE40-AS1 |
BHLHE40 antisense RNA 1 |
26290 |
0.17 |
| chr18_2884812_2884995 | 0.33 |
RP11-737O24.5 |
|
36061 |
0.12 |
| chr7_110650904_110651055 | 0.32 |
LRRN3 |
leucine rich repeat neuronal 3 |
80083 |
0.1 |
| chr19_2701835_2702122 | 0.32 |
GNG7 |
guanine nucleotide binding protein (G protein), gamma 7 |
729 |
0.53 |
| chr9_36666940_36667091 | 0.32 |
MELK |
maternal embryonic leucine zipper kinase |
94107 |
0.08 |
| chr20_47365648_47365892 | 0.32 |
ENSG00000251876 |
. |
9785 |
0.28 |
| chr2_234150632_234150905 | 0.32 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
9449 |
0.15 |
| chr2_43382252_43382437 | 0.32 |
ENSG00000207087 |
. |
63712 |
0.12 |
| chr8_144075541_144075692 | 0.32 |
LY6E |
lymphocyte antigen 6 complex, locus E |
23783 |
0.15 |
| chr16_23464322_23464473 | 0.32 |
COG7 |
component of oligomeric golgi complex 7 |
104 |
0.95 |
| chrX_147701487_147701638 | 0.32 |
AFF2-IT1 |
AFF2 intronic transcript 1 (non-protein coding) |
73163 |
0.11 |
| chr19_12936277_12936463 | 0.32 |
CTD-2265O21.3 |
|
155 |
0.86 |
| chr1_161097729_161097968 | 0.32 |
DEDD |
death effector domain containing |
2779 |
0.1 |
| chr20_19918143_19918294 | 0.32 |
RIN2 |
Ras and Rab interactor 2 |
48008 |
0.15 |
| chr3_52331644_52331795 | 0.32 |
ENSG00000242797 |
. |
1211 |
0.27 |
| chr2_111610296_111610719 | 0.31 |
ACOXL |
acyl-CoA oxidase-like |
47611 |
0.18 |
| chr6_157304957_157305108 | 0.31 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
82525 |
0.11 |
| chr14_89925929_89926200 | 0.31 |
FOXN3 |
forkhead box N3 |
34331 |
0.15 |
| chr19_30557968_30558119 | 0.31 |
URI1 |
URI1, prefoldin-like chaperone |
54713 |
0.17 |
| chr12_117534939_117535195 | 0.31 |
TESC |
tescalcin |
2184 |
0.41 |
| chr16_3554613_3554882 | 0.31 |
CLUAP1 |
clusterin associated protein 1 |
2703 |
0.18 |
| chr7_143005618_143005769 | 0.31 |
CLCN1 |
chloride channel, voltage-sensitive 1 |
7526 |
0.1 |
| chr14_107198967_107199211 | 0.31 |
IGHV3-72 |
immunoglobulin heavy variable 3-72 |
382 |
0.58 |
| chr14_23018397_23018864 | 0.31 |
AE000662.92 |
Uncharacterized protein |
6904 |
0.11 |
| chr2_98347856_98348007 | 0.31 |
ZAP70 |
zeta-chain (TCR) associated protein kinase 70kDa |
2938 |
0.25 |
| chr6_167508030_167508365 | 0.31 |
CCR6 |
chemokine (C-C motif) receptor 6 |
17098 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.4 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.2 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.2 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 0.3 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.1 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.2 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.1 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.5 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.5 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.2 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.2 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0021780 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process(GO:0009129) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:2000328 | T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) memory T cell differentiation(GO:0043379) regulation of memory T cell differentiation(GO:0043380) positive regulation of memory T cell differentiation(GO:0043382) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 17 cell differentiation(GO:2000319) positive regulation of T-helper 17 cell differentiation(GO:2000321) regulation of T-helper 17 cell lineage commitment(GO:2000328) positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.2 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.2 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.0 | 0.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0031065 | regulation of histone deacetylation(GO:0031063) positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:1904019 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.2 | GO:0044409 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.0 | GO:0060685 | regulation of prostatic bud formation(GO:0060685) |
| 0.0 | 0.1 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:2000192 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0032367 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.0 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0051461 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0045446 | endothelial cell differentiation(GO:0045446) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.6 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.1 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.2 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.2 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.3 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.0 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.7 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.0 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |