Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
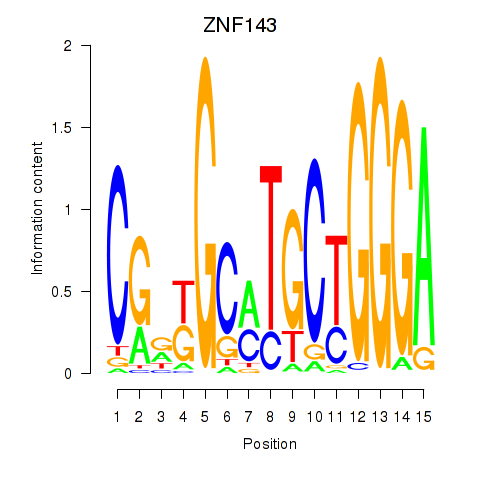
Results for ZNF143
Z-value: 1.65
Transcription factors associated with ZNF143
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF143
|
ENSG00000166478.5 | zinc finger protein 143 |
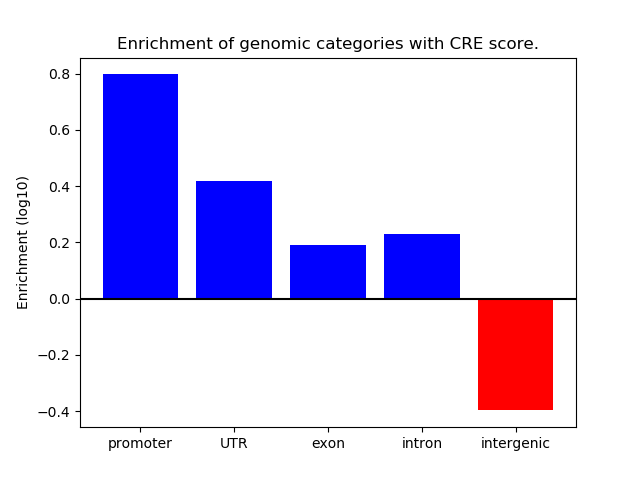
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_9511208_9511359 | ZNF143 | 22761 | 0.140938 | 0.79 | 1.1e-02 | Click! |
| chr11_9481615_9481873 | ZNF143 | 122 | 0.691450 | -0.79 | 1.1e-02 | Click! |
| chr11_9556875_9557026 | ZNF143 | 22906 | 0.138514 | 0.77 | 1.4e-02 | Click! |
| chr11_9510681_9510832 | ZNF143 | 23288 | 0.139671 | 0.71 | 3.4e-02 | Click! |
| chr11_9532170_9532321 | ZNF143 | 1799 | 0.327001 | -0.66 | 5.5e-02 | Click! |
Activity of the ZNF143 motif across conditions
Conditions sorted by the z-value of the ZNF143 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
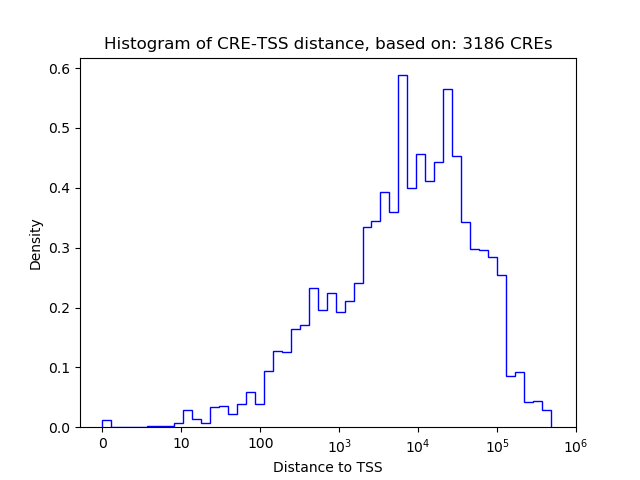
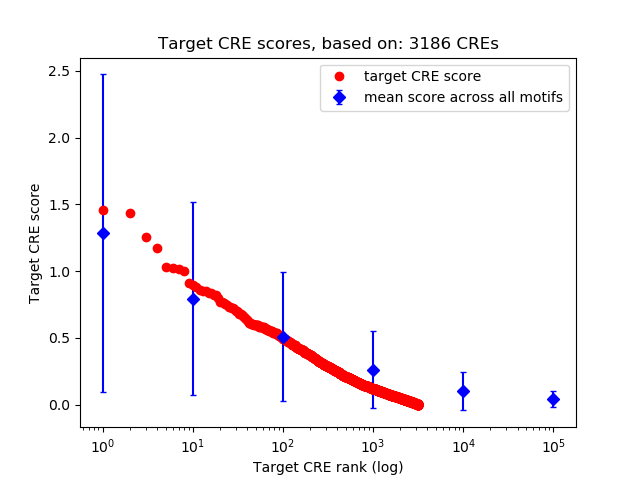
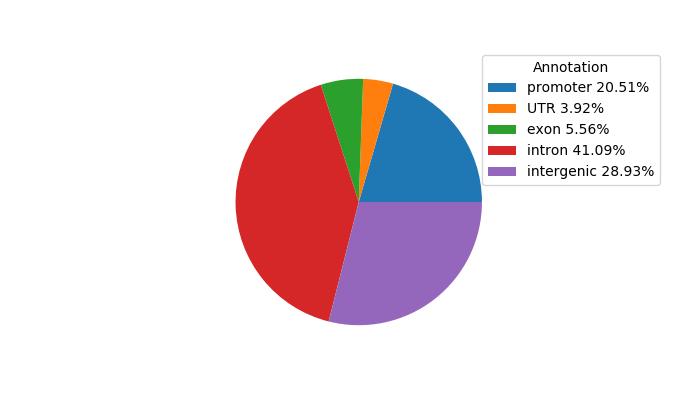
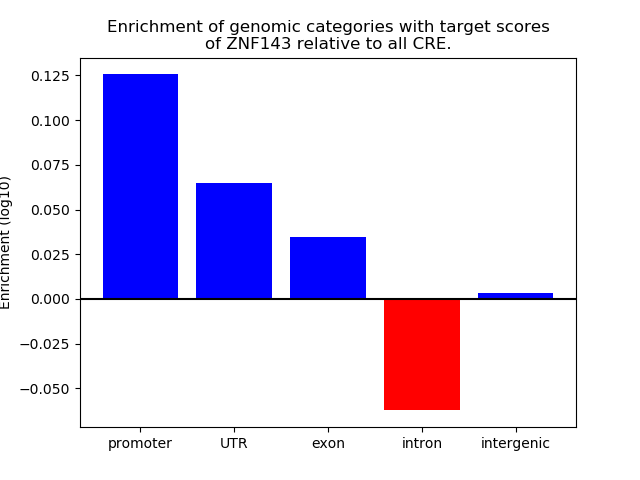
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_41912974_41913725 | 1.46 |
MGA |
MGA, MAX dimerization protein |
385 |
0.85 |
| chr1_150510557_150510858 | 1.43 |
ADAMTSL4 |
ADAMTS-like 4 |
11177 |
0.08 |
| chr9_132236336_132236633 | 1.25 |
ENSG00000264298 |
. |
4351 |
0.26 |
| chr8_37556598_37557288 | 1.17 |
ZNF703 |
zinc finger protein 703 |
3674 |
0.16 |
| chr8_49341467_49342503 | 1.03 |
ENSG00000252710 |
. |
121395 |
0.06 |
| chr16_85424724_85424875 | 1.02 |
RP11-680G10.1 |
Uncharacterized protein |
33730 |
0.18 |
| chr18_21464195_21464346 | 1.02 |
LAMA3 |
laminin, alpha 3 |
467 |
0.84 |
| chr1_8272810_8272977 | 1.00 |
ENSG00000200975 |
. |
6236 |
0.24 |
| chr18_42179529_42179680 | 0.91 |
SETBP1 |
SET binding protein 1 |
80534 |
0.12 |
| chr16_88366428_88366870 | 0.89 |
ZNF469 |
zinc finger protein 469 |
127230 |
0.05 |
| chr10_104385999_104386279 | 0.88 |
TRIM8 |
tripartite motif containing 8 |
18114 |
0.15 |
| chr16_56884007_56884388 | 0.86 |
ENSG00000207649 |
. |
8233 |
0.14 |
| chr21_17959428_17959579 | 0.85 |
ENSG00000207863 |
. |
3054 |
0.31 |
| chr2_238804572_238804786 | 0.85 |
ENSG00000263723 |
. |
26132 |
0.17 |
| chr2_43301822_43302026 | 0.83 |
ENSG00000207087 |
. |
16708 |
0.28 |
| chr2_19340414_19340714 | 0.83 |
ENSG00000266738 |
. |
207626 |
0.03 |
| chr1_203524952_203525250 | 0.83 |
OPTC |
opticin |
60003 |
0.11 |
| chr17_14821652_14821803 | 0.82 |
ENSG00000238806 |
. |
319473 |
0.01 |
| chr17_36604180_36604649 | 0.80 |
ENSG00000260833 |
. |
3518 |
0.2 |
| chr11_35551231_35551382 | 0.77 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
542 |
0.83 |
| chr1_32038621_32038861 | 0.77 |
RP11-73M7.1 |
|
2433 |
0.21 |
| chr11_72500407_72500904 | 0.76 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
3643 |
0.14 |
| chr9_130817852_130818146 | 0.76 |
RP11-379C10.1 |
|
1642 |
0.22 |
| chr9_37966595_37966746 | 0.75 |
ENSG00000251745 |
. |
29905 |
0.19 |
| chr1_17908500_17908722 | 0.73 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
1563 |
0.51 |
| chr3_138581340_138581807 | 0.73 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
27793 |
0.2 |
| chr13_110161938_110162089 | 0.72 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
218616 |
0.02 |
| chr21_37583097_37583248 | 0.72 |
ENSG00000265882 |
. |
2964 |
0.22 |
| chr14_23316976_23317711 | 0.71 |
ENSG00000212335 |
. |
4594 |
0.09 |
| chr17_9331018_9331169 | 0.70 |
AC087501.1 |
|
24421 |
0.18 |
| chr1_230379381_230379663 | 0.70 |
RP5-956O18.2 |
|
24707 |
0.21 |
| chr15_48919832_48919983 | 0.70 |
FBN1 |
fibrillin 1 |
18011 |
0.26 |
| chr17_17614010_17614161 | 0.68 |
RAI1 |
retinoic acid induced 1 |
28276 |
0.14 |
| chr3_177234511_177234662 | 0.68 |
ENSG00000252028 |
. |
13246 |
0.3 |
| chr10_73019589_73019740 | 0.67 |
UNC5B-AS1 |
UNC5B antisense RNA 1 |
41679 |
0.12 |
| chr3_52080953_52081104 | 0.66 |
DUSP7 |
dual specificity phosphatase 7 |
9153 |
0.12 |
| chr5_168587659_168587810 | 0.66 |
ENSG00000207619 |
. |
102964 |
0.07 |
| chr11_61055093_61055244 | 0.65 |
VWCE |
von Willebrand factor C and EGF domains |
7728 |
0.13 |
| chr11_57123702_57124132 | 0.64 |
ENSG00000266018 |
. |
3279 |
0.12 |
| chr17_55952277_55952762 | 0.64 |
CUEDC1 |
CUE domain containing 1 |
2488 |
0.26 |
| chr11_65670447_65670677 | 0.63 |
FOSL1 |
FOS-like antigen 1 |
2518 |
0.12 |
| chr16_56883616_56883799 | 0.61 |
ENSG00000207649 |
. |
8723 |
0.14 |
| chr8_38539787_38539938 | 0.61 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
45842 |
0.14 |
| chr10_111841442_111842330 | 0.61 |
ADD3 |
adducin 3 (gamma) |
74164 |
0.09 |
| chr19_48903661_48904050 | 0.61 |
GRIN2D |
glutamate receptor, ionotropic, N-methyl D-aspartate 2D |
5723 |
0.11 |
| chr1_117028094_117028245 | 0.61 |
ENSG00000200547 |
. |
33011 |
0.13 |
| chr20_36772405_36772603 | 0.60 |
TGM2 |
transglutaminase 2 |
21168 |
0.17 |
| chr10_81200859_81201010 | 0.60 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
3041 |
0.28 |
| chr13_43819793_43820081 | 0.60 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
115276 |
0.07 |
| chr22_46849624_46849775 | 0.60 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
81492 |
0.08 |
| chr7_48290426_48290608 | 0.60 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
79460 |
0.11 |
| chr8_99376791_99376994 | 0.60 |
KCNS2 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
62358 |
0.11 |
| chr1_17909315_17909466 | 0.59 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
2342 |
0.4 |
| chr3_182880353_182880504 | 0.59 |
LAMP3 |
lysosomal-associated membrane protein 3 |
242 |
0.94 |
| chr3_8649116_8649267 | 0.59 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
4395 |
0.22 |
| chr17_74382503_74382654 | 0.58 |
SPHK1 |
sphingosine kinase 1 |
1205 |
0.31 |
| chr6_14540959_14541110 | 0.58 |
ENSG00000206960 |
. |
105732 |
0.08 |
| chr11_10373135_10373286 | 0.58 |
AMPD3 |
adenosine monophosphate deaminase 3 |
33992 |
0.13 |
| chr13_28902618_28902769 | 0.58 |
FLT1 |
fms-related tyrosine kinase 1 |
5923 |
0.29 |
| chr17_17669783_17670067 | 0.58 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
4210 |
0.15 |
| chr19_46876400_46876551 | 0.58 |
AC007193.9 |
|
9951 |
0.12 |
| chr6_128828631_128828782 | 0.57 |
RP1-86D1.4 |
|
2436 |
0.25 |
| chr16_49734250_49734401 | 0.57 |
ENSG00000221134 |
. |
21194 |
0.22 |
| chr16_72961357_72961552 | 0.57 |
ENSG00000221799 |
. |
57302 |
0.12 |
| chr7_5452523_5452740 | 0.57 |
TNRC18 |
trinucleotide repeat containing 18 |
5606 |
0.15 |
| chr15_80109103_80109367 | 0.56 |
ENSG00000252995 |
. |
557 |
0.82 |
| chr6_14743458_14743640 | 0.56 |
ENSG00000206960 |
. |
96783 |
0.09 |
| chr17_36667174_36667728 | 0.56 |
ARHGAP23 |
Rho GTPase activating protein 23 |
13416 |
0.16 |
| chr1_43918298_43918577 | 0.56 |
HYI |
hydroxypyruvate isomerase (putative) |
686 |
0.48 |
| chr10_72056262_72056676 | 0.56 |
NPFFR1 |
neuropeptide FF receptor 1 |
13037 |
0.21 |
| chr7_93976913_93977064 | 0.55 |
COL1A2 |
collagen, type I, alpha 2 |
46885 |
0.18 |
| chr2_218574918_218575069 | 0.55 |
DIRC3 |
disrupted in renal carcinoma 3 |
46285 |
0.18 |
| chr2_239773492_239773709 | 0.55 |
TWIST2 |
twist family bHLH transcription factor 2 |
16927 |
0.24 |
| chr14_75919638_75919789 | 0.55 |
JDP2 |
Jun dimerization protein 2 |
20876 |
0.17 |
| chr21_44918501_44918652 | 0.55 |
SIK1 |
salt-inducible kinase 1 |
71568 |
0.11 |
| chr15_67316146_67316685 | 0.54 |
SMAD3 |
SMAD family member 3 |
39686 |
0.2 |
| chr7_90794057_90794258 | 0.54 |
FZD1 |
frizzled family receptor 1 |
99626 |
0.09 |
| chr17_2310691_2310842 | 0.54 |
MNT |
MAX network transcriptional repressor |
6354 |
0.12 |
| chr15_67457558_67457709 | 0.54 |
SMAD3 |
SMAD family member 3 |
724 |
0.72 |
| chr17_25681100_25681558 | 0.54 |
WSB1 |
WD repeat and SOCS box containing 1 |
59957 |
0.11 |
| chr1_201645041_201645675 | 0.54 |
RP11-25B7.1 |
|
2086 |
0.26 |
| chr1_27057613_27057764 | 0.54 |
ARID1A |
AT rich interactive domain 1A (SWI-like) |
1817 |
0.28 |
| chr6_46111105_46111256 | 0.53 |
ENPP4 |
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative) |
13450 |
0.18 |
| chr9_113309012_113309163 | 0.53 |
ENSG00000212254 |
. |
7009 |
0.26 |
| chr19_49004091_49004498 | 0.53 |
LMTK3 |
lemur tyrosine kinase 3 |
10767 |
0.1 |
| chr16_70726438_70726684 | 0.53 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
2935 |
0.19 |
| chr12_49663177_49663419 | 0.53 |
RP11-161H23.5 |
|
3791 |
0.13 |
| chr20_5100313_5100464 | 0.52 |
PCNA |
proliferating cell nuclear antigen |
284 |
0.83 |
| chr17_79393744_79394123 | 0.52 |
RP11-1055B8.7 |
BAH and coiled-coil domain-containing protein 1 |
11463 |
0.11 |
| chr8_49676509_49676660 | 0.52 |
EFCAB1 |
EF-hand calcium binding domain 1 |
28714 |
0.25 |
| chr19_52233260_52233541 | 0.52 |
HAS1 |
hyaluronan synthase 1 |
6153 |
0.11 |
| chr1_68252004_68252155 | 0.52 |
ENSG00000238778 |
. |
13743 |
0.21 |
| chr12_76052830_76052981 | 0.51 |
ENSG00000251893 |
. |
30179 |
0.2 |
| chr14_75926083_75926288 | 0.51 |
JDP2 |
Jun dimerization protein 2 |
27348 |
0.16 |
| chr21_47813743_47813894 | 0.51 |
PCNT |
pericentrin |
43136 |
0.11 |
| chr15_58733197_58733348 | 0.51 |
LIPC |
lipase, hepatic |
9082 |
0.21 |
| chr8_22598834_22598985 | 0.51 |
ENSG00000253125 |
. |
6107 |
0.18 |
| chr7_116331971_116332122 | 0.51 |
MET |
met proto-oncogene |
7093 |
0.26 |
| chr2_95987573_95987724 | 0.50 |
KCNIP3 |
Kv channel interacting protein 3, calsenilin |
11678 |
0.23 |
| chr8_145734382_145734533 | 0.50 |
MFSD3 |
major facilitator superfamily domain containing 3 |
0 |
0.92 |
| chr7_8149373_8149524 | 0.49 |
AC006042.6 |
|
4207 |
0.27 |
| chr2_241861602_241861753 | 0.49 |
AC104809.3 |
Protein LOC728763 |
309 |
0.87 |
| chr16_84844160_84844846 | 0.49 |
CRISPLD2 |
cysteine-rich secretory protein LCCL domain containing 2 |
9087 |
0.18 |
| chr16_88366119_88366347 | 0.49 |
ZNF469 |
zinc finger protein 469 |
127646 |
0.05 |
| chr1_201992311_201992462 | 0.49 |
RP11-510N19.5 |
|
11888 |
0.11 |
| chr2_171381606_171381757 | 0.49 |
AC007277.3 |
|
118773 |
0.06 |
| chr12_49580416_49580658 | 0.49 |
TUBA1A |
tubulin, alpha 1a |
110 |
0.95 |
| chr5_14055268_14055419 | 0.49 |
TRIO |
trio Rho guanine nucleotide exchange factor |
88468 |
0.1 |
| chr22_24870211_24870362 | 0.48 |
UPB1 |
ureidopropionase, beta |
7080 |
0.16 |
| chr16_57180947_57181162 | 0.48 |
FAM192A |
family with sequence similarity 192, member A |
26646 |
0.12 |
| chr6_148271708_148271859 | 0.48 |
SASH1 |
SAM and SH3 domain containing 1 |
321657 |
0.01 |
| chr3_54939961_54940112 | 0.48 |
CACNA2D3-AS1 |
CACNA2D3 antisense RNA 1 |
4754 |
0.29 |
| chr7_18809339_18809490 | 0.48 |
ENSG00000222164 |
. |
38488 |
0.21 |
| chr17_54851727_54851878 | 0.47 |
C17orf67 |
chromosome 17 open reading frame 67 |
41448 |
0.14 |
| chr11_73309592_73309967 | 0.47 |
FAM168A |
family with sequence similarity 168, member A |
545 |
0.79 |
| chr1_231917552_231917705 | 0.47 |
ENSG00000222986 |
. |
111247 |
0.06 |
| chr11_133995520_133995672 | 0.47 |
NCAPD3 |
non-SMC condensin II complex, subunit D3 |
42771 |
0.13 |
| chr12_112621415_112621566 | 0.47 |
HECTD4 |
HECT domain containing E3 ubiquitin protein ligase 4 |
6984 |
0.18 |
| chr9_115940859_115941010 | 0.47 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
27712 |
0.15 |
| chr6_8180536_8180832 | 0.47 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
77873 |
0.11 |
| chr1_17636370_17636667 | 0.46 |
PADI4 |
peptidyl arginine deiminase, type IV |
1826 |
0.31 |
| chr12_130646297_130646669 | 0.46 |
FZD10 |
frizzled family receptor 10 |
521 |
0.88 |
| chr20_62089406_62089557 | 0.46 |
RP11-358D14.2 |
|
10077 |
0.12 |
| chr4_186651858_186652115 | 0.46 |
SORBS2 |
sorbin and SH3 domain containing 2 |
9498 |
0.21 |
| chr22_39688804_39689038 | 0.46 |
RP3-333H23.8 |
|
72 |
0.95 |
| chr20_1783720_1783871 | 0.45 |
SIRPA |
signal-regulatory protein alpha |
91359 |
0.07 |
| chr7_93539808_93539959 | 0.45 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
3977 |
0.19 |
| chr11_114248963_114249341 | 0.45 |
RP11-64D24.4 |
|
1615 |
0.32 |
| chr8_97279888_97280058 | 0.45 |
PTDSS1 |
phosphatidylserine synthase 1 |
5832 |
0.17 |
| chr3_42693962_42694141 | 0.45 |
ZBTB47 |
zinc finger and BTB domain containing 47 |
1125 |
0.31 |
| chr5_102780433_102780701 | 0.45 |
NUDT12 |
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
117923 |
0.06 |
| chr2_99528935_99529086 | 0.45 |
KIAA1211L |
KIAA1211-like |
4026 |
0.3 |
| chr1_155223556_155224415 | 0.45 |
FAM189B |
family with sequence similarity 189, member B |
714 |
0.38 |
| chr5_54280930_54281081 | 0.44 |
ESM1 |
endothelial cell-specific molecule 1 |
416 |
0.82 |
| chr19_2529730_2529881 | 0.44 |
ENSG00000252962 |
. |
26666 |
0.13 |
| chr5_177963455_177963606 | 0.44 |
COL23A1 |
collagen, type XXIII, alpha 1 |
25709 |
0.2 |
| chr7_101457741_101458764 | 0.44 |
CUX1 |
cut-like homeobox 1 |
707 |
0.73 |
| chr12_125327332_125327720 | 0.44 |
SCARB1 |
scavenger receptor class B, member 1 |
2937 |
0.3 |
| chr5_151054870_151055036 | 0.43 |
CTB-113P19.1 |
|
1553 |
0.32 |
| chr2_217970592_217970881 | 0.43 |
ENSG00000251849 |
. |
70485 |
0.12 |
| chr4_16028808_16029222 | 0.43 |
ENSG00000251758 |
. |
18605 |
0.2 |
| chr1_17951607_17951758 | 0.43 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
6833 |
0.29 |
| chr15_45748380_45748570 | 0.43 |
RP11-519G16.3 |
|
2733 |
0.15 |
| chr8_11373209_11373360 | 0.43 |
BLK |
B lymphoid tyrosine kinase |
21385 |
0.13 |
| chr11_59524240_59524533 | 0.43 |
STX3 |
syntaxin 3 |
1461 |
0.29 |
| chr16_70716878_70717029 | 0.43 |
MTSS1L |
metastasis suppressor 1-like |
3016 |
0.19 |
| chr17_75417410_75417842 | 0.43 |
SEPT9 |
septin 9 |
1742 |
0.31 |
| chr5_176914993_176915144 | 0.43 |
RP11-1334A24.6 |
|
6928 |
0.1 |
| chrX_19621349_19621543 | 0.42 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
67034 |
0.13 |
| chr12_93516624_93516849 | 0.42 |
RP11-511B23.2 |
|
16131 |
0.2 |
| chr19_18571206_18571357 | 0.42 |
ELL |
elongation factor RNA polymerase II |
15462 |
0.09 |
| chr17_2310928_2311079 | 0.42 |
MNT |
MAX network transcriptional repressor |
6591 |
0.12 |
| chr22_36903062_36903213 | 0.42 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
11 |
0.97 |
| chr5_1004113_1004264 | 0.42 |
ENSG00000221244 |
. |
114 |
0.95 |
| chr16_152002_152153 | 0.42 |
Z69720.2 |
|
11448 |
0.09 |
| chr1_45279408_45279559 | 0.42 |
BTBD19 |
BTB (POZ) domain containing 19 |
5288 |
0.08 |
| chr12_25959266_25959417 | 0.42 |
ENSG00000222950 |
. |
26838 |
0.24 |
| chr7_41926654_41927141 | 0.41 |
AC005027.3 |
|
181964 |
0.03 |
| chr19_8017646_8018209 | 0.41 |
CTD-3193O13.14 |
|
6080 |
0.08 |
| chr3_124826438_124826589 | 0.41 |
ENSG00000199327 |
. |
11531 |
0.17 |
| chr7_137621728_137621986 | 0.41 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
3221 |
0.27 |
| chr2_28625085_28625236 | 0.41 |
RP11-373D23.2 |
|
5478 |
0.18 |
| chr17_38643211_38643393 | 0.41 |
TNS4 |
tensin 4 |
8402 |
0.16 |
| chr9_124650254_124650616 | 0.41 |
RP11-244O19.1 |
|
68704 |
0.12 |
| chr19_6220741_6220892 | 0.41 |
RFX2 |
regulatory factor X, 2 (influences HLA class II expression) |
21276 |
0.14 |
| chr11_94523601_94523752 | 0.41 |
AMOTL1 |
angiomotin like 1 |
22139 |
0.21 |
| chr9_96354377_96354528 | 0.41 |
PHF2 |
PHD finger protein 2 |
15543 |
0.19 |
| chr20_2852821_2853294 | 0.41 |
PTPRA |
protein tyrosine phosphatase, receptor type, A |
1009 |
0.46 |
| chr20_359199_359350 | 0.40 |
TRIB3 |
tribbles pseudokinase 3 |
1987 |
0.25 |
| chr10_99332980_99333131 | 0.40 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
519 |
0.68 |
| chr3_194892357_194892562 | 0.40 |
XXYLT1 |
xyloside xylosyltransferase 1 |
3943 |
0.18 |
| chr9_138749633_138749784 | 0.40 |
CAMSAP1 |
calmodulin regulated spectrin-associated protein 1 |
25035 |
0.18 |
| chr19_9473586_9473877 | 0.39 |
ZNF177 |
zinc finger protein 177 |
24 |
0.97 |
| chr12_49983022_49983173 | 0.39 |
ENSG00000199237 |
. |
3790 |
0.15 |
| chr22_36278822_36279066 | 0.39 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
42310 |
0.19 |
| chr12_19728777_19728928 | 0.39 |
AEBP2 |
AE binding protein 2 |
75789 |
0.11 |
| chr12_13025678_13025838 | 0.39 |
GPRC5A |
G protein-coupled receptor, family C, group 5, member A |
17958 |
0.14 |
| chr10_73625407_73625558 | 0.39 |
PSAP |
prosaposin |
14356 |
0.19 |
| chr1_112275365_112275766 | 0.39 |
FAM212B |
family with sequence similarity 212, member B |
6310 |
0.18 |
| chr11_65075775_65076171 | 0.39 |
CDC42EP2 |
CDC42 effector protein (Rho GTPase binding) 2 |
6316 |
0.13 |
| chr12_15754427_15754578 | 0.39 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
20805 |
0.23 |
| chr15_74277001_74277287 | 0.39 |
STOML1 |
stomatin (EPB72)-like 1 |
7031 |
0.16 |
| chr18_33887353_33887504 | 0.39 |
FHOD3 |
formin homology 2 domain containing 3 |
9629 |
0.28 |
| chr7_40611624_40611778 | 0.38 |
AC004988.1 |
|
25174 |
0.27 |
| chr13_24782132_24782457 | 0.38 |
SPATA13 |
spermatogenesis associated 13 |
43538 |
0.13 |
| chr14_76447358_76447593 | 0.38 |
TGFB3 |
transforming growth factor, beta 3 |
59 |
0.98 |
| chr17_76993895_76994046 | 0.38 |
CANT1 |
calcium activated nucleotidase 1 |
11842 |
0.13 |
| chr6_150246933_150247085 | 0.38 |
RAET1G |
retinoic acid early transcript 1G |
2752 |
0.18 |
| chr20_60944733_60944974 | 0.38 |
LAMA5 |
laminin, alpha 5 |
2485 |
0.18 |
| chr2_9777860_9778099 | 0.38 |
YWHAQ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
6836 |
0.21 |
| chr5_398631_398914 | 0.38 |
AHRR |
aryl-hydrocarbon receptor repressor |
22263 |
0.14 |
| chrX_20085662_20085960 | 0.38 |
MAP7D2 |
MAP7 domain containing 2 |
10916 |
0.18 |
| chr20_48450027_48450178 | 0.38 |
ENSG00000252123 |
. |
5573 |
0.18 |
| chr13_40105757_40105908 | 0.38 |
LHFP |
lipoma HMGIC fusion partner |
71476 |
0.11 |
| chr22_30841104_30841316 | 0.37 |
MTFP1 |
mitochondrial fission process 1 |
19405 |
0.07 |
| chr19_16177822_16178237 | 0.37 |
TPM4 |
tropomyosin 4 |
198 |
0.94 |
| chr22_45933959_45934110 | 0.37 |
FBLN1 |
fibulin 1 |
19371 |
0.2 |
| chr10_95226291_95226450 | 0.37 |
MYOF |
myoferlin |
15581 |
0.19 |
| chr13_97959313_97959591 | 0.37 |
MBNL2 |
muscleblind-like splicing regulator 2 |
30994 |
0.21 |
| chr19_46524126_46524277 | 0.37 |
ENSG00000211580 |
. |
2011 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.3 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.2 | GO:0046084 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.1 | 0.2 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.1 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.1 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.2 | GO:0071883 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.2 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.3 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.2 | GO:0033604 | negative regulation of catecholamine secretion(GO:0033604) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0033240 | positive regulation of cellular amine metabolic process(GO:0033240) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.7 | GO:0034333 | adherens junction assembly(GO:0034333) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0090505 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly(GO:0090504) epiboly involved in wound healing(GO:0090505) |
| 0.0 | 0.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.2 | GO:1901072 | amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.6 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:2000846 | corticosteroid hormone secretion(GO:0035930) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.4 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) ERAD pathway(GO:0036503) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.1 | GO:0034644 | cellular response to UV(GO:0034644) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.3 | GO:0019674 | NAD metabolic process(GO:0019674) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0070977 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.0 | GO:0044557 | relaxation of smooth muscle(GO:0044557) negative regulation of smooth muscle contraction(GO:0045986) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0042462 | photoreceptor cell development(GO:0042461) eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0033598 | mammary gland epithelial cell proliferation(GO:0033598) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.0 | GO:0010659 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.0 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.2 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.0 | 0.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.4 | GO:0051084 | 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.1 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 1.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 0.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.1 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.0 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 1.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |