Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for ZNF148
Z-value: 0.81
Transcription factors associated with ZNF148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF148
|
ENSG00000163848.14 | zinc finger protein 148 |
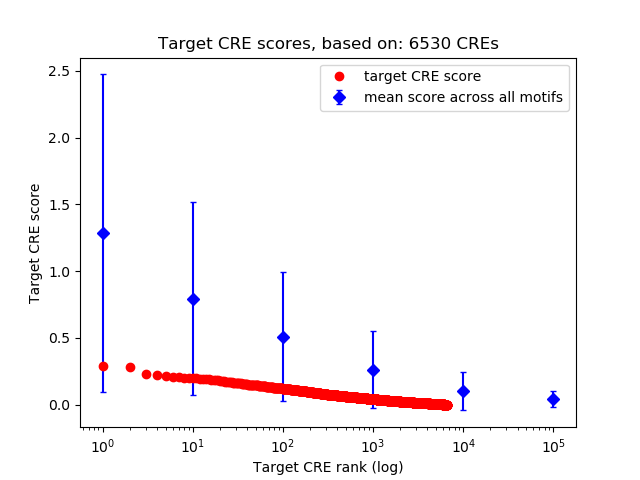
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_125089012_125089267 | ZNF148 | 4954 | 0.222347 | 0.71 | 3.1e-02 | Click! |
| chr3_125055856_125056007 | ZNF148 | 20193 | 0.190897 | 0.67 | 4.6e-02 | Click! |
| chr3_125074033_125074184 | ZNF148 | 2016 | 0.351729 | 0.55 | 1.3e-01 | Click! |
| chr3_125081016_125081167 | ZNF148 | 4429 | 0.235327 | 0.51 | 1.6e-01 | Click! |
| chr3_125073758_125073909 | ZNF148 | 2291 | 0.323920 | 0.49 | 1.8e-01 | Click! |
Activity of the ZNF148 motif across conditions
Conditions sorted by the z-value of the ZNF148 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
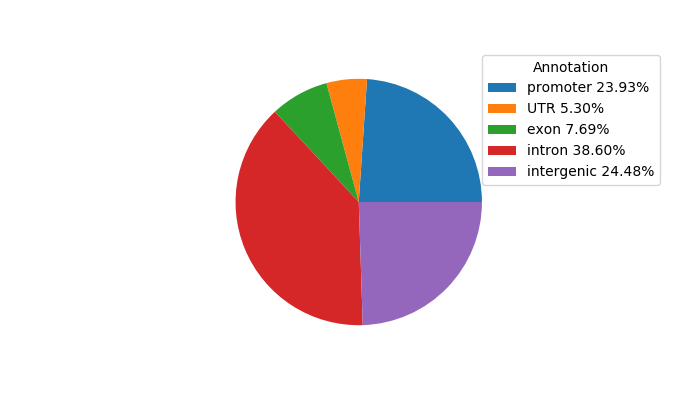
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_15328141_15328292 | 0.29 |
KAZN |
kazrin, periplakin interacting protein |
55801 |
0.16 |
| chr1_23850956_23851507 | 0.28 |
E2F2 |
E2F transcription factor 2 |
6481 |
0.19 |
| chr3_47027623_47027925 | 0.23 |
CCDC12 |
coiled-coil domain containing 12 |
4302 |
0.2 |
| chr22_47253139_47253290 | 0.22 |
ENSG00000221672 |
. |
9411 |
0.26 |
| chr10_6573467_6573756 | 0.21 |
PRKCQ |
protein kinase C, theta |
48590 |
0.18 |
| chr12_112608315_112608466 | 0.21 |
HECTD4 |
HECT domain containing E3 ubiquitin protein ligase 4 |
6116 |
0.17 |
| chr22_40588636_40588787 | 0.21 |
TNRC6B |
trinucleotide repeat containing 6B |
14765 |
0.25 |
| chr1_5450304_5450501 | 0.20 |
ENSG00000264341 |
. |
173729 |
0.04 |
| chr1_9781158_9781391 | 0.20 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
3147 |
0.23 |
| chr6_36684328_36684602 | 0.20 |
RAB44 |
RAB44, member RAS oncogene family |
1209 |
0.43 |
| chr14_93364438_93364589 | 0.20 |
CHGA |
chromogranin A (parathyroid secretory protein 1) |
24912 |
0.22 |
| chr11_115630994_115631145 | 0.19 |
ENSG00000239153 |
. |
133166 |
0.06 |
| chr9_130541716_130542127 | 0.19 |
SH2D3C |
SH2 domain containing 3C |
901 |
0.36 |
| chr13_26465091_26465379 | 0.19 |
AL138815.2 |
Uncharacterized protein |
12556 |
0.2 |
| chr22_32602732_32602927 | 0.19 |
RP1-90G24.10 |
|
1727 |
0.27 |
| chr1_111180164_111180315 | 0.19 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
6143 |
0.19 |
| chr11_63729654_63729805 | 0.18 |
ENSG00000207200 |
. |
8213 |
0.11 |
| chr17_38261346_38261675 | 0.18 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
4532 |
0.13 |
| chr20_37491189_37491977 | 0.18 |
ENSG00000240474 |
. |
9830 |
0.2 |
| chr19_4389183_4389334 | 0.18 |
SH3GL1 |
SH3-domain GRB2-like 1 |
1349 |
0.22 |
| chr5_175461916_175462142 | 0.18 |
THOC3 |
THO complex 3 |
346 |
0.84 |
| chr10_6079008_6079159 | 0.18 |
IL2RA |
interleukin 2 receptor, alpha |
11119 |
0.14 |
| chr15_38831010_38831161 | 0.17 |
ENSG00000201509 |
. |
3359 |
0.22 |
| chr19_40448721_40449473 | 0.17 |
FCGBP |
Fc fragment of IgG binding protein |
8564 |
0.16 |
| chr17_2705001_2705152 | 0.17 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
5300 |
0.2 |
| chr6_36630670_36630821 | 0.17 |
ENSG00000251864 |
. |
9870 |
0.14 |
| chr1_903446_903597 | 0.17 |
PLEKHN1 |
pleckstrin homology domain containing, family N member 1 |
1639 |
0.19 |
| chr8_27182129_27182563 | 0.16 |
PTK2B |
protein tyrosine kinase 2 beta |
516 |
0.81 |
| chr2_99123851_99124123 | 0.16 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
12525 |
0.24 |
| chr17_48993495_48993783 | 0.16 |
TOB1 |
transducer of ERBB2, 1 |
48300 |
0.11 |
| chr10_126315803_126316294 | 0.16 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
76146 |
0.09 |
| chr10_133999367_134000256 | 0.16 |
RP11-140A10.3 |
|
345 |
0.67 |
| chr5_177236057_177236208 | 0.16 |
FAM153A |
family with sequence similarity 153, member A |
25733 |
0.14 |
| chr2_106423458_106423752 | 0.16 |
NCK2 |
NCK adaptor protein 2 |
9410 |
0.27 |
| chr17_76113070_76113221 | 0.16 |
ENSG00000204282 |
. |
5265 |
0.13 |
| chr15_31670738_31670889 | 0.15 |
KLF13 |
Kruppel-like factor 13 |
12456 |
0.3 |
| chr11_118746193_118746372 | 0.15 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
8193 |
0.1 |
| chr22_40058850_40059001 | 0.15 |
CACNA1I |
calcium channel, voltage-dependent, T type, alpha 1I subunit |
92167 |
0.07 |
| chr3_134406340_134406491 | 0.15 |
KY |
kyphoscoliosis peptidase |
36551 |
0.18 |
| chr22_38614803_38615828 | 0.15 |
MAFF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
5773 |
0.12 |
| chr11_134526662_134526813 | 0.15 |
B3GAT1 |
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
244925 |
0.02 |
| chr19_38909040_38909370 | 0.15 |
RASGRP4 |
RAS guanyl releasing protein 4 |
7597 |
0.09 |
| chr1_6479268_6479610 | 0.15 |
HES2 |
hes family bHLH transcription factor 2 |
524 |
0.65 |
| chr19_1073799_1074502 | 0.15 |
HMHA1 |
histocompatibility (minor) HA-1 |
2638 |
0.13 |
| chr15_61107075_61107226 | 0.15 |
RP11-554D20.1 |
|
50211 |
0.16 |
| chr9_138981963_138982279 | 0.15 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
5010 |
0.23 |
| chr20_48528466_48528617 | 0.15 |
SPATA2 |
spermatogenesis associated 2 |
1735 |
0.28 |
| chr9_96342392_96342543 | 0.15 |
PHF2 |
PHD finger protein 2 |
3558 |
0.24 |
| chr11_119560225_119560376 | 0.15 |
ENSG00000199217 |
. |
33279 |
0.14 |
| chr19_13280797_13280977 | 0.15 |
CTC-250I14.6 |
|
15880 |
0.1 |
| chr11_866339_866490 | 0.15 |
RP11-1391J7.1 |
|
6619 |
0.08 |
| chr1_211509622_211509899 | 0.15 |
TRAF5 |
TNF receptor-associated factor 5 |
9581 |
0.25 |
| chr12_133064825_133065258 | 0.14 |
FBRSL1 |
fibrosin-like 1 |
1096 |
0.54 |
| chr4_1234799_1234950 | 0.14 |
CTBP1 |
C-terminal binding protein 1 |
837 |
0.53 |
| chr12_106691482_106691807 | 0.14 |
TCP11L2 |
t-complex 11, testis-specific-like 2 |
4063 |
0.2 |
| chr1_154386815_154386966 | 0.14 |
RP11-350G8.5 |
|
7850 |
0.13 |
| chr4_7940034_7940471 | 0.14 |
AC097381.1 |
|
476 |
0.75 |
| chr1_18993186_18993337 | 0.14 |
PAX7 |
paired box 7 |
35243 |
0.21 |
| chr1_203260343_203260644 | 0.14 |
BTG2 |
BTG family, member 2 |
14171 |
0.17 |
| chr19_41113680_41113831 | 0.14 |
ENSG00000266164 |
. |
800 |
0.53 |
| chr10_82012672_82012823 | 0.14 |
AL359195.1 |
Uncharacterized protein; cDNA FLJ46261 fis, clone TESTI4025062 |
3281 |
0.25 |
| chr19_13206082_13206442 | 0.14 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
7419 |
0.1 |
| chr15_89195219_89195432 | 0.14 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
13141 |
0.17 |
| chr10_30347671_30348341 | 0.14 |
KIAA1462 |
KIAA1462 |
447 |
0.9 |
| chr13_40945604_40945755 | 0.14 |
ENSG00000252812 |
. |
35898 |
0.22 |
| chr7_156837851_156838002 | 0.14 |
MNX1-AS1 |
MNX1 antisense RNA 1 (head to head) |
34427 |
0.13 |
| chr12_122214020_122214201 | 0.14 |
RHOF |
ras homolog family member F (in filopodia) |
17156 |
0.15 |
| chr6_14636111_14636367 | 0.14 |
ENSG00000206960 |
. |
10527 |
0.32 |
| chr19_11689000_11689260 | 0.13 |
ACP5 |
acid phosphatase 5, tartrate resistant |
179 |
0.91 |
| chr8_145048593_145049035 | 0.13 |
PLEC |
plectin |
723 |
0.48 |
| chr17_26876127_26876878 | 0.13 |
UNC119 |
unc-119 homolog (C. elegans) |
85 |
0.93 |
| chr1_12014718_12014869 | 0.13 |
PLOD1 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
19983 |
0.11 |
| chr10_135236057_135236208 | 0.13 |
SPRN |
shadow of prion protein homolog (zebrafish) |
1944 |
0.2 |
| chr6_112352232_112352383 | 0.13 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
22968 |
0.19 |
| chr15_61080818_61080969 | 0.13 |
RP11-554D20.1 |
|
23954 |
0.22 |
| chr22_32598678_32598909 | 0.13 |
RFPL2 |
ret finger protein-like 2 |
671 |
0.62 |
| chr13_113420676_113420874 | 0.13 |
ATP11A-AS1 |
ATP11A antisense RNA 1 |
11768 |
0.19 |
| chr1_27176686_27176837 | 0.13 |
ZDHHC18 |
zinc finger, DHHC-type containing 18 |
92 |
0.94 |
| chr9_139689207_139689358 | 0.13 |
CCDC183 |
coiled-coil domain containing 183 |
1533 |
0.16 |
| chr10_125846427_125846578 | 0.13 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
5468 |
0.31 |
| chr9_80635334_80635485 | 0.13 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
10111 |
0.31 |
| chr19_3176335_3176657 | 0.13 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
2240 |
0.2 |
| chr2_240873719_240873870 | 0.13 |
ENSG00000264279 |
. |
8717 |
0.24 |
| chr1_203294414_203294865 | 0.13 |
ENSG00000202300 |
. |
6412 |
0.18 |
| chr1_162291884_162292035 | 0.13 |
RP11-565P22.2 |
|
4375 |
0.15 |
| chr8_134304078_134304332 | 0.13 |
NDRG1 |
N-myc downstream regulated 1 |
4574 |
0.3 |
| chr13_32616590_32616779 | 0.12 |
FRY-AS1 |
FRY antisense RNA 1 |
10908 |
0.22 |
| chr12_104295444_104295595 | 0.12 |
HSP90B1 |
heat shock protein 90kDa beta (Grp94), member 1 |
28366 |
0.12 |
| chr18_12307599_12307896 | 0.12 |
TUBB6 |
tubulin, beta 6 class V |
79 |
0.97 |
| chr9_92686685_92686836 | 0.12 |
ENSG00000263967 |
. |
99057 |
0.09 |
| chr17_80061984_80062253 | 0.12 |
CCDC57 |
coiled-coil domain containing 57 |
2392 |
0.14 |
| chr2_219158817_219159019 | 0.12 |
TMBIM1 |
transmembrane BAX inhibitor motif containing 1 |
1609 |
0.23 |
| chr11_35148064_35148393 | 0.12 |
AL356215.1 |
|
1987 |
0.3 |
| chr17_76774924_76775177 | 0.12 |
CYTH1 |
cytohesin 1 |
3304 |
0.24 |
| chr16_57021435_57021712 | 0.12 |
NLRC5 |
NLR family, CARD domain containing 5 |
1837 |
0.25 |
| chr11_118742943_118743241 | 0.12 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
11383 |
0.1 |
| chr11_3923915_3924127 | 0.12 |
STIM1 |
stromal interaction molecule 1 |
672 |
0.62 |
| chr11_63975572_63976297 | 0.12 |
FERMT3 |
fermitin family member 3 |
645 |
0.44 |
| chrX_153671917_153672203 | 0.12 |
FAM50A |
family with sequence similarity 50, member A |
413 |
0.63 |
| chr13_114801042_114801193 | 0.12 |
RASA3 |
RAS p21 protein activator 3 |
42321 |
0.18 |
| chr8_134184001_134184169 | 0.12 |
WISP1 |
WNT1 inducible signaling pathway protein 1 |
19197 |
0.2 |
| chr7_156838975_156839126 | 0.12 |
MNX1-AS1 |
MNX1 antisense RNA 1 (head to head) |
35551 |
0.13 |
| chr19_12896860_12897296 | 0.12 |
ENSG00000263800 |
. |
864 |
0.31 |
| chr9_92050627_92050944 | 0.12 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
593 |
0.81 |
| chr5_139283800_139284387 | 0.12 |
NRG2 |
neuregulin 2 |
111 |
0.97 |
| chr9_134609016_134609403 | 0.12 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
3716 |
0.25 |
| chr8_142413099_142413596 | 0.12 |
CTD-3064M3.4 |
|
10673 |
0.12 |
| chr15_58775556_58775753 | 0.12 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
9542 |
0.18 |
| chr11_64619085_64619254 | 0.12 |
CDC42BPG |
CDC42 binding protein kinase gamma (DMPK-like) |
7128 |
0.11 |
| chr1_14066743_14066960 | 0.12 |
PRDM2 |
PR domain containing 2, with ZNF domain |
9047 |
0.24 |
| chr19_46283041_46283318 | 0.12 |
DMPK |
dystrophia myotonica-protein kinase |
620 |
0.48 |
| chr2_234142703_234142912 | 0.12 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
17410 |
0.14 |
| chr17_48262095_48262525 | 0.12 |
COL1A1 |
collagen, type I, alpha 1 |
15242 |
0.09 |
| chr1_26686481_26686807 | 0.12 |
AIM1L |
absent in melanoma 1-like |
6023 |
0.12 |
| chr19_17907446_17907815 | 0.12 |
B3GNT3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
1664 |
0.26 |
| chr5_169259346_169259518 | 0.12 |
CTB-37A13.1 |
|
53063 |
0.15 |
| chr1_200364105_200364256 | 0.12 |
ZNF281 |
zinc finger protein 281 |
14940 |
0.29 |
| chr1_59408762_59408913 | 0.12 |
JUN |
jun proto-oncogene |
159052 |
0.04 |
| chr1_203246394_203246639 | 0.12 |
BTG2 |
BTG family, member 2 |
28148 |
0.14 |
| chr19_12949497_12949966 | 0.12 |
MAST1 |
microtubule associated serine/threonine kinase 1 |
383 |
0.64 |
| chr1_110046721_110046872 | 0.11 |
AMIGO1 |
adhesion molecule with Ig-like domain 1 |
5508 |
0.11 |
| chr19_2043146_2043605 | 0.11 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
743 |
0.49 |
| chr22_49307613_49307764 | 0.11 |
ENSG00000251912 |
. |
97472 |
0.09 |
| chr19_41196829_41197444 | 0.11 |
NUMBL |
numb homolog (Drosophila)-like |
259 |
0.87 |
| chr2_231120036_231120187 | 0.11 |
SP140 |
SP140 nuclear body protein |
29632 |
0.16 |
| chr4_991996_992147 | 0.11 |
IDUA |
iduronidase, alpha-L- |
1427 |
0.3 |
| chr11_64692978_64693146 | 0.11 |
PPP2R5B |
protein phosphatase 2, regulatory subunit B', beta |
869 |
0.39 |
| chr5_180586986_180587137 | 0.11 |
OR2V2 |
olfactory receptor, family 2, subfamily V, member 2 |
5118 |
0.12 |
| chr8_22435891_22436512 | 0.11 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
53 |
0.95 |
| chr3_177272692_177272963 | 0.11 |
ENSG00000252028 |
. |
51487 |
0.17 |
| chr16_50634708_50634859 | 0.11 |
RP11-401P9.6 |
|
12812 |
0.16 |
| chr11_65621570_65621754 | 0.11 |
CFL1 |
cofilin 1 (non-muscle) |
2282 |
0.13 |
| chrX_135718612_135718763 | 0.11 |
ENSG00000233093 |
. |
3015 |
0.22 |
| chr22_20819732_20820026 | 0.11 |
ENSG00000255156 |
. |
9719 |
0.1 |
| chr6_159194328_159194479 | 0.11 |
ENSG00000265558 |
. |
8618 |
0.2 |
| chr3_13526745_13527110 | 0.11 |
HDAC11 |
histone deacetylase 11 |
4181 |
0.22 |
| chr19_1017328_1017803 | 0.11 |
TMEM259 |
transmembrane protein 259 |
143 |
0.89 |
| chr11_128555893_128556312 | 0.11 |
RP11-744N12.3 |
|
221 |
0.62 |
| chrX_48546553_48546795 | 0.11 |
WAS |
Wiskott-Aldrich syndrome |
4506 |
0.14 |
| chr2_68942118_68942311 | 0.11 |
ARHGAP25 |
Rho GTPase activating protein 25 |
4580 |
0.29 |
| chr19_1789686_1789853 | 0.11 |
ATP8B3 |
ATPase, aminophospholipid transporter, class I, type 8B, member 3 |
21854 |
0.09 |
| chr15_32656988_32657139 | 0.11 |
ENSG00000221444 |
. |
28610 |
0.11 |
| chr17_43315078_43315229 | 0.11 |
FMNL1 |
formin-like 1 |
3289 |
0.12 |
| chr22_39487886_39488384 | 0.11 |
RP4-742C19.12 |
|
544 |
0.68 |
| chr19_39896092_39896528 | 0.11 |
ZFP36 |
ZFP36 ring finger protein |
1143 |
0.25 |
| chr17_38287853_38288004 | 0.11 |
MSL1 |
male-specific lethal 1 homolog (Drosophila) |
7098 |
0.12 |
| chr10_134069250_134069918 | 0.11 |
STK32C |
serine/threonine kinase 32C |
51134 |
0.12 |
| chr2_99253969_99254120 | 0.11 |
UNC50 |
unc-50 homolog (C. elegans) |
21375 |
0.17 |
| chr2_106792435_106792586 | 0.11 |
AC018878.3 |
|
3488 |
0.29 |
| chr14_76032303_76032574 | 0.11 |
FLVCR2 |
feline leukemia virus subgroup C cellular receptor family, member 2 |
12522 |
0.16 |
| chr22_19862093_19862244 | 0.11 |
GNB1L |
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
19706 |
0.14 |
| chr17_4439765_4439919 | 0.11 |
SPNS2 |
spinster homolog 2 (Drosophila) |
3515 |
0.15 |
| chr18_46333522_46333736 | 0.11 |
RP11-484L8.1 |
|
27512 |
0.22 |
| chr2_31207015_31207166 | 0.11 |
AC009305.1 |
|
2715 |
0.36 |
| chr9_139419925_139420076 | 0.10 |
ENSG00000263403 |
. |
5922 |
0.1 |
| chr1_206872496_206872728 | 0.10 |
MAPKAPK2 |
mitogen-activated protein kinase-activated protein kinase 2 |
14230 |
0.14 |
| chr11_119592281_119592574 | 0.10 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
6836 |
0.21 |
| chr11_60756317_60756468 | 0.10 |
ENSG00000207153 |
. |
3753 |
0.15 |
| chr10_126343736_126343887 | 0.10 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
48383 |
0.14 |
| chr1_21979132_21979283 | 0.10 |
RAP1GAP |
RAP1 GTPase activating protein |
593 |
0.77 |
| chrX_135776880_135777031 | 0.10 |
CD40LG |
CD40 ligand |
46569 |
0.11 |
| chr5_1502002_1502255 | 0.10 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
21964 |
0.2 |
| chr17_75457461_75457675 | 0.10 |
SEPT9 |
septin 9 |
4917 |
0.17 |
| chr2_9554076_9554227 | 0.10 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
6272 |
0.21 |
| chr1_25229799_25229950 | 0.10 |
RUNX3 |
runt-related transcription factor 3 |
25738 |
0.2 |
| chr7_1572770_1573329 | 0.10 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
2699 |
0.2 |
| chr16_88367012_88367357 | 0.10 |
ZNF469 |
zinc finger protein 469 |
126695 |
0.05 |
| chr1_85354747_85355038 | 0.10 |
LPAR3 |
lysophosphatidic acid receptor 3 |
4004 |
0.29 |
| chr20_60892338_60892489 | 0.10 |
RP11-157P1.4 |
|
11032 |
0.12 |
| chr7_100463554_100463705 | 0.10 |
TRIP6 |
thyroid hormone receptor interactor 6 |
1131 |
0.3 |
| chr15_52758556_52758733 | 0.10 |
MYO5A |
myosin VA (heavy chain 12, myoxin) |
30777 |
0.2 |
| chr19_13954343_13954494 | 0.10 |
ENSG00000207980 |
. |
6945 |
0.08 |
| chr1_156456694_156456845 | 0.10 |
ENSG00000206651 |
. |
2767 |
0.17 |
| chr17_76128893_76129294 | 0.10 |
TMC6 |
transmembrane channel-like 6 |
605 |
0.58 |
| chr11_121262801_121262980 | 0.10 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
60022 |
0.13 |
| chr19_35748150_35748384 | 0.10 |
AC002128.5 |
|
3725 |
0.11 |
| chr9_92455695_92455846 | 0.10 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
235817 |
0.02 |
| chr17_33192377_33192528 | 0.10 |
CCT6B |
chaperonin containing TCP1, subunit 6B (zeta 2) |
95969 |
0.06 |
| chr11_64623671_64624265 | 0.10 |
CDC42BPG |
CDC42 binding protein kinase gamma (DMPK-like) |
11927 |
0.1 |
| chr5_1524609_1524892 | 0.10 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
658 |
0.76 |
| chr1_172677197_172677413 | 0.10 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
49147 |
0.17 |
| chr1_147015470_147015621 | 0.10 |
BCL9 |
B-cell CLL/lymphoma 9 |
2363 |
0.39 |
| chr17_18943804_18943955 | 0.10 |
GRAP |
GRB2-related adaptor protein |
1919 |
0.21 |
| chr20_49258194_49258345 | 0.10 |
RP4-530I15.6 |
|
3739 |
0.18 |
| chr9_95800059_95800489 | 0.10 |
SUSD3 |
sushi domain containing 3 |
20715 |
0.16 |
| chr22_40009284_40009473 | 0.10 |
CACNA1I |
calcium channel, voltage-dependent, T type, alpha 1I subunit |
42620 |
0.14 |
| chr20_32053414_32053565 | 0.10 |
ENSG00000264923 |
. |
6635 |
0.17 |
| chr7_75624660_75624811 | 0.10 |
POR |
P450 (cytochrome) oxidoreductase |
13506 |
0.15 |
| chr14_24783456_24784225 | 0.10 |
LTB4R |
leukotriene B4 receptor |
66 |
0.91 |
| chr20_30298367_30298518 | 0.10 |
AL160175.1 |
|
10559 |
0.13 |
| chr2_239072757_239072908 | 0.10 |
FAM132B |
family with sequence similarity 132, member B |
142 |
0.94 |
| chr11_405167_405318 | 0.10 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
1128 |
0.25 |
| chr1_228074508_228074659 | 0.10 |
ENSG00000264483 |
. |
54801 |
0.1 |
| chr14_21556036_21556544 | 0.10 |
ZNF219 |
zinc finger protein 219 |
5697 |
0.1 |
| chr4_153858873_153859306 | 0.10 |
FHDC1 |
FH2 domain containing 1 |
1585 |
0.47 |
| chr17_66380201_66380484 | 0.10 |
ENSG00000207561 |
. |
40347 |
0.14 |
| chr1_161097323_161097680 | 0.10 |
DEDD |
death effector domain containing |
3126 |
0.09 |
| chr9_116310824_116310999 | 0.10 |
RGS3 |
regulator of G-protein signaling 3 |
12127 |
0.22 |
| chr21_16582633_16582784 | 0.09 |
NRIP1 |
nuclear receptor interacting protein 1 |
145387 |
0.05 |
| chr6_166744143_166744377 | 0.09 |
SFT2D1 |
SFT2 domain containing 1 |
11819 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0003094 | regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine production(GO:0032682) negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0002251 | organ or tissue specific immune response(GO:0002251) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.0 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:2000189 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.0 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |