Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
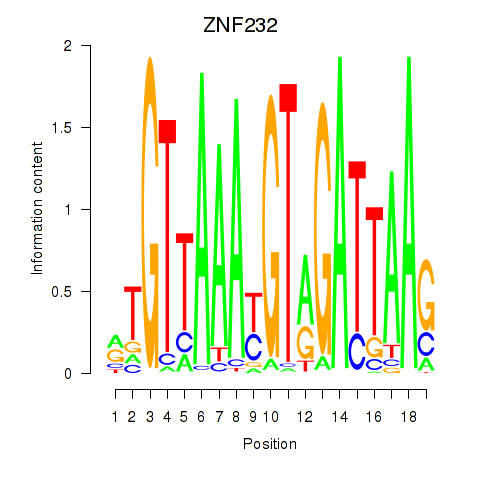
Results for ZNF232
Z-value: 1.13
Transcription factors associated with ZNF232
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF232
|
ENSG00000167840.9 | zinc finger protein 232 |
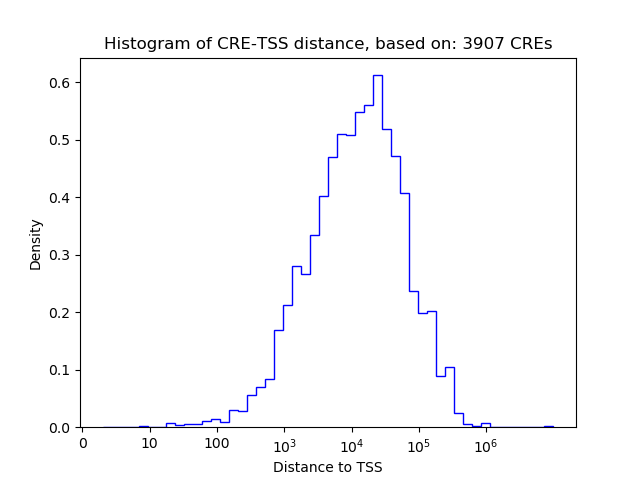
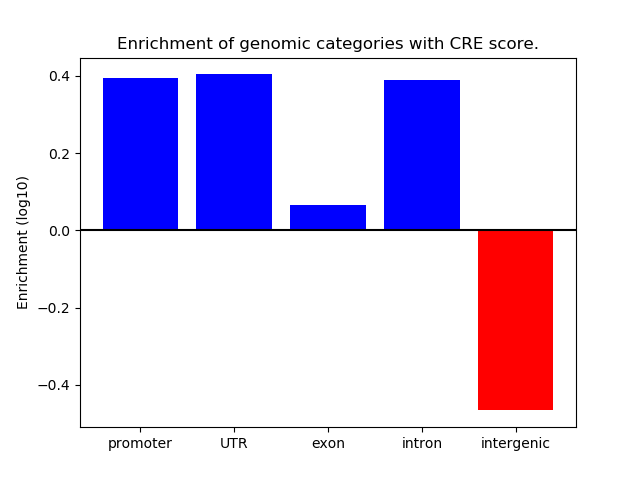
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_5014893_5015044 | ZNF232 | 168 | 0.531362 | -0.84 | 4.3e-03 | Click! |
| chr17_5025884_5026035 | ZNF232 | 438 | 0.711549 | -0.46 | 2.2e-01 | Click! |
| chr17_5028739_5028890 | ZNF232 | 2417 | 0.161305 | -0.38 | 3.1e-01 | Click! |
| chr17_5025598_5025749 | ZNF232 | 724 | 0.508643 | -0.38 | 3.2e-01 | Click! |
| chr17_5026950_5027101 | ZNF232 | 628 | 0.568930 | -0.31 | 4.2e-01 | Click! |
Activity of the ZNF232 motif across conditions
Conditions sorted by the z-value of the ZNF232 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
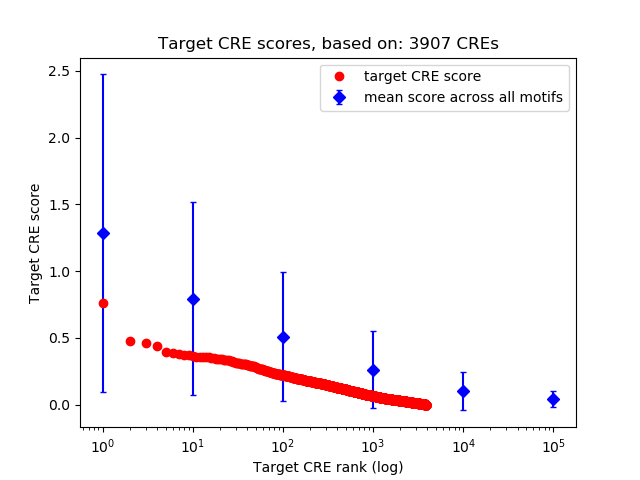
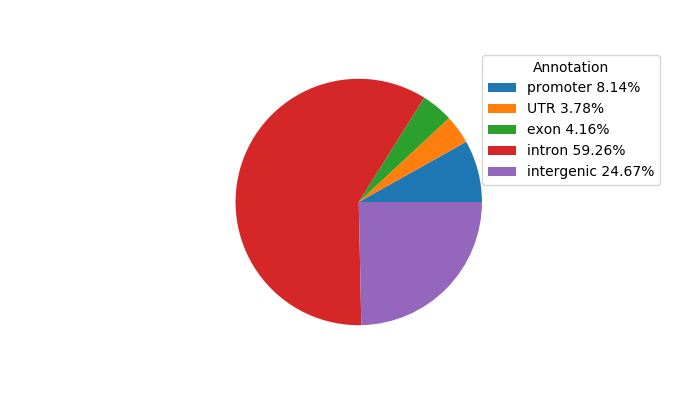
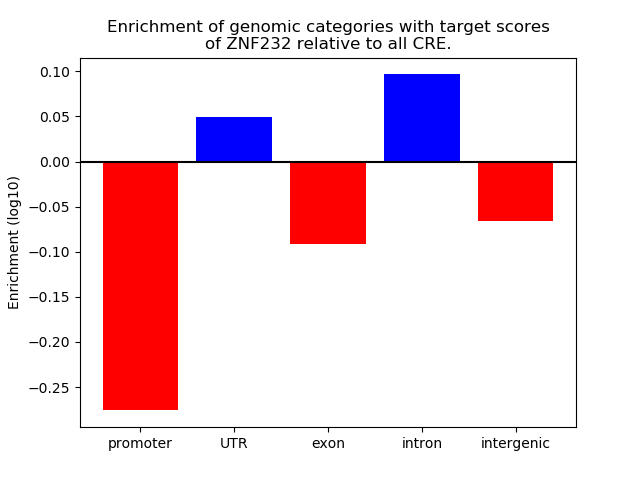
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_108557025_108557176 | 0.76 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
15481 |
0.25 |
| chr3_38071955_38072561 | 0.48 |
PLCD1 |
phospholipase C, delta 1 |
1005 |
0.45 |
| chr10_33422454_33423119 | 0.46 |
ENSG00000263576 |
. |
35222 |
0.17 |
| chr13_94982893_94983044 | 0.44 |
ENSG00000212057 |
. |
134676 |
0.05 |
| chr12_124145145_124145296 | 0.39 |
TCTN2 |
tectonic family member 2 |
10440 |
0.13 |
| chr10_31631628_31631809 | 0.39 |
RP11-192P3.5 |
|
21219 |
0.17 |
| chr3_4864499_4864721 | 0.38 |
ENSG00000239126 |
. |
55776 |
0.13 |
| chr18_67557274_67557425 | 0.37 |
CD226 |
CD226 molecule |
57306 |
0.16 |
| chr6_128068941_128069092 | 0.37 |
THEMIS |
thymocyte selection associated |
153087 |
0.04 |
| chr4_186133064_186133215 | 0.36 |
SNX25 |
sorting nexin 25 |
1906 |
0.3 |
| chr6_139483644_139483926 | 0.36 |
HECA |
headcase homolog (Drosophila) |
27536 |
0.2 |
| chr2_69815178_69815329 | 0.36 |
AAK1 |
AP2 associated kinase 1 |
55533 |
0.11 |
| chr13_98113590_98113741 | 0.36 |
RAP2A |
RAP2A, member of RAS oncogene family |
27189 |
0.24 |
| chr1_90077786_90078098 | 0.36 |
RP11-413E1.4 |
|
17342 |
0.16 |
| chr1_121293995_121294146 | 0.35 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
358133 |
0.01 |
| chr14_64340904_64341076 | 0.35 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
21258 |
0.2 |
| chr5_66456559_66456909 | 0.35 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
18472 |
0.24 |
| chr3_150167839_150167990 | 0.34 |
TSC22D2 |
TSC22 domain family, member 2 |
39118 |
0.18 |
| chr7_28164005_28164156 | 0.34 |
ENSG00000206623 |
. |
6242 |
0.26 |
| chr22_20909677_20910043 | 0.34 |
MED15 |
mediator complex subunit 15 |
4283 |
0.17 |
| chr13_50830244_50830433 | 0.34 |
ENSG00000221198 |
. |
107035 |
0.07 |
| chr9_71991121_71991272 | 0.34 |
FAM189A2 |
family with sequence similarity 189, member A2 |
5014 |
0.31 |
| chr8_37097408_37097594 | 0.33 |
RP11-150O12.6 |
|
277038 |
0.01 |
| chr1_194022845_194023072 | 0.33 |
ENSG00000252241 |
. |
321884 |
0.01 |
| chr10_33225735_33225886 | 0.33 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
631 |
0.82 |
| chr8_67443611_67443762 | 0.33 |
ENSG00000206949 |
. |
30140 |
0.15 |
| chr13_74899245_74899396 | 0.33 |
ENSG00000206617 |
. |
35969 |
0.21 |
| chr3_66330811_66331009 | 0.32 |
ENSG00000206759 |
. |
12734 |
0.22 |
| chr10_90763076_90763284 | 0.32 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
12033 |
0.14 |
| chr2_219089063_219089333 | 0.31 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
7265 |
0.12 |
| chr6_112121057_112121208 | 0.31 |
FYN |
FYN oncogene related to SRC, FGR, YES |
1506 |
0.54 |
| chr2_106459836_106459987 | 0.31 |
NCK2 |
NCK adaptor protein 2 |
8293 |
0.24 |
| chr12_20657108_20657259 | 0.31 |
RP11-284H19.1 |
|
133987 |
0.05 |
| chr11_104786300_104786451 | 0.31 |
RP11-693N9.2 |
|
5733 |
0.21 |
| chr10_33315822_33316332 | 0.31 |
ENSG00000265319 |
. |
7775 |
0.23 |
| chr2_26048696_26048925 | 0.31 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
45632 |
0.16 |
| chr6_161498090_161498241 | 0.31 |
MAP3K4 |
mitogen-activated protein kinase kinase kinase 4 |
54939 |
0.14 |
| chr22_23132033_23132342 | 0.30 |
ENSG00000207833 |
. |
2790 |
0.07 |
| chr1_78143107_78143258 | 0.30 |
ZZZ3 |
zinc finger, ZZ-type containing 3 |
5161 |
0.21 |
| chr2_205736509_205736732 | 0.30 |
PARD3B |
par-3 family cell polarity regulator beta |
325897 |
0.01 |
| chr2_106707234_106707385 | 0.30 |
C2orf40 |
chromosome 2 open reading frame 40 |
25053 |
0.21 |
| chr3_43079553_43079704 | 0.29 |
FAM198A |
family with sequence similarity 198, member A |
58588 |
0.1 |
| chr15_60684276_60684517 | 0.29 |
ANXA2 |
annexin A2 |
900 |
0.69 |
| chr6_143203048_143203338 | 0.29 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
45009 |
0.18 |
| chr17_47736485_47736636 | 0.29 |
SPOP |
speckle-type POZ protein |
12568 |
0.15 |
| chr17_13971675_13971896 | 0.29 |
ENSG00000236088 |
. |
990 |
0.42 |
| chr18_43737463_43737614 | 0.29 |
C18orf25 |
chromosome 18 open reading frame 25 |
15981 |
0.17 |
| chr5_141485413_141485564 | 0.29 |
NDFIP1 |
Nedd4 family interacting protein 1 |
2582 |
0.34 |
| chr3_124722968_124723119 | 0.28 |
HEG1 |
heart development protein with EGF-like domains 1 |
2179 |
0.34 |
| chr12_96598172_96598568 | 0.28 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
9978 |
0.2 |
| chr12_47003400_47003570 | 0.28 |
SLC38A4 |
solute carrier family 38, member 4 |
159602 |
0.04 |
| chr12_4401971_4402452 | 0.27 |
CCND2-AS1 |
CCND2 antisense RNA 1 |
16861 |
0.15 |
| chr3_109619904_109620055 | 0.27 |
ENSG00000265956 |
. |
298304 |
0.01 |
| chr5_49698412_49698563 | 0.27 |
EMB |
embigin |
26074 |
0.28 |
| chr6_158182790_158182947 | 0.27 |
SNX9 |
sorting nexin 9 |
61428 |
0.12 |
| chr7_114597355_114597535 | 0.27 |
MDFIC |
MyoD family inhibitor domain containing |
23521 |
0.28 |
| chr7_95130919_95131189 | 0.27 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
15841 |
0.18 |
| chr2_55125265_55125416 | 0.27 |
ENSG00000252507 |
. |
47914 |
0.15 |
| chr7_6292541_6292692 | 0.27 |
CYTH3 |
cytohesin 3 |
19659 |
0.16 |
| chr20_3137233_3137451 | 0.26 |
UBOX5 |
U-box domain containing 5 |
3180 |
0.15 |
| chr4_109513219_109513370 | 0.26 |
ENSG00000266046 |
. |
14998 |
0.2 |
| chr3_45666862_45667013 | 0.26 |
LIMD1 |
LIM domains containing 1 |
30586 |
0.15 |
| chr3_160159792_160159943 | 0.26 |
TRIM59 |
tripartite motif containing 59 |
6858 |
0.15 |
| chrX_10076834_10077577 | 0.26 |
WWC3 |
WWC family member 3 |
45706 |
0.12 |
| chr18_7963851_7964002 | 0.25 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
16974 |
0.22 |
| chr1_40914947_40915118 | 0.25 |
ZFP69B |
ZFP69 zinc finger protein B |
747 |
0.56 |
| chr6_16432796_16433123 | 0.25 |
ENSG00000265642 |
. |
4205 |
0.35 |
| chr8_125630679_125630830 | 0.25 |
RP11-532M24.1 |
|
38882 |
0.15 |
| chr7_15502538_15502689 | 0.25 |
AGMO |
alkylglycerol monooxygenase |
96766 |
0.09 |
| chr11_64640724_64641020 | 0.25 |
EHD1 |
EH-domain containing 1 |
2266 |
0.16 |
| chr6_136200607_136200783 | 0.25 |
PDE7B |
phosphodiesterase 7B |
27861 |
0.2 |
| chrX_46472704_46472855 | 0.24 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
39560 |
0.17 |
| chr3_55639839_55639990 | 0.24 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
115941 |
0.07 |
| chr2_12410968_12411119 | 0.24 |
ENSG00000264089 |
. |
71787 |
0.13 |
| chr11_96006440_96006591 | 0.24 |
ENSG00000266192 |
. |
68087 |
0.11 |
| chr1_222594825_222595030 | 0.24 |
ENSG00000222399 |
. |
82150 |
0.1 |
| chr10_27514870_27515021 | 0.24 |
ENSG00000251839 |
. |
6300 |
0.14 |
| chr5_80294021_80294254 | 0.24 |
CTC-459I6.1 |
|
37411 |
0.17 |
| chr8_69645584_69645735 | 0.23 |
ENSG00000239184 |
. |
37781 |
0.22 |
| chr2_213996401_213996552 | 0.23 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
16877 |
0.28 |
| chr2_239347338_239347489 | 0.23 |
AC016999.2 |
|
11043 |
0.17 |
| chr10_15183836_15183987 | 0.23 |
NMT2 |
N-myristoyltransferase 2 |
18213 |
0.17 |
| chr6_75657197_75657348 | 0.23 |
COL12A1 |
collagen, type XII, alpha 1 |
171502 |
0.03 |
| chr4_80918707_80918911 | 0.23 |
ANTXR2 |
anthrax toxin receptor 2 |
74908 |
0.12 |
| chr13_98573793_98573944 | 0.23 |
IPO5 |
importin 5 |
32044 |
0.2 |
| chr9_4689321_4689472 | 0.23 |
CDC37L1 |
cell division cycle 37-like 1 |
9830 |
0.17 |
| chr6_144880670_144880834 | 0.23 |
UTRN |
utrophin |
23594 |
0.28 |
| chr15_93523585_93523736 | 0.23 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
8547 |
0.26 |
| chr6_112063218_112063369 | 0.23 |
FYN |
FYN oncogene related to SRC, FGR, YES |
17024 |
0.24 |
| chr12_51702385_51702551 | 0.23 |
BIN2 |
bridging integrator 2 |
15431 |
0.14 |
| chr14_99726600_99726784 | 0.23 |
AL109767.1 |
|
2593 |
0.31 |
| chr6_169891020_169891171 | 0.23 |
WDR27 |
WD repeat domain 27 |
169693 |
0.03 |
| chr15_78443266_78443417 | 0.23 |
IDH3A |
isocitrate dehydrogenase 3 (NAD+) alpha |
1608 |
0.31 |
| chr4_38686540_38686749 | 0.23 |
RP11-617D20.1 |
|
20140 |
0.15 |
| chr13_22025148_22025299 | 0.22 |
ZDHHC20 |
zinc finger, DHHC-type containing 20 |
8191 |
0.22 |
| chr2_169290019_169290449 | 0.22 |
ENSG00000239230 |
. |
7481 |
0.21 |
| chr2_128120808_128120959 | 0.22 |
MAP3K2 |
mitogen-activated protein kinase kinase kinase 2 |
20078 |
0.16 |
| chrY_2720348_2720499 | 0.22 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
10462 |
0.24 |
| chr1_29272903_29273060 | 0.22 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
31890 |
0.15 |
| chr7_114595751_114595902 | 0.22 |
MDFIC |
MyoD family inhibitor domain containing |
21902 |
0.28 |
| chr10_131741460_131741611 | 0.22 |
EBF3 |
early B-cell factor 3 |
20570 |
0.27 |
| chr15_52951493_52951915 | 0.22 |
FAM214A |
family with sequence similarity 214, member A |
7457 |
0.21 |
| chr15_77749022_77749295 | 0.22 |
HMG20A |
high mobility group 20A |
1401 |
0.51 |
| chr1_24021474_24021706 | 0.22 |
RPL11 |
ribosomal protein L11 |
2491 |
0.23 |
| chr3_108323323_108323612 | 0.22 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
1769 |
0.36 |
| chr3_190099541_190099692 | 0.22 |
CLDN16 |
claudin 16 |
6045 |
0.26 |
| chr1_117914498_117914649 | 0.22 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
4502 |
0.33 |
| chr2_182089447_182089783 | 0.22 |
ENSG00000266705 |
. |
80764 |
0.11 |
| chr10_91393117_91393268 | 0.22 |
PANK1 |
pantothenate kinase 1 |
10439 |
0.18 |
| chr9_74375290_74375441 | 0.22 |
TMEM2 |
transmembrane protein 2 |
7937 |
0.3 |
| chr11_61311867_61312018 | 0.22 |
LRRC10B |
leucine rich repeat containing 10B |
35670 |
0.11 |
| chr18_46464398_46464549 | 0.21 |
SMAD7 |
SMAD family member 7 |
10402 |
0.25 |
| chr1_46281591_46281742 | 0.21 |
MAST2 |
microtubule associated serine/threonine kinase 2 |
12381 |
0.22 |
| chr22_22121001_22121154 | 0.21 |
ENSG00000200985 |
. |
25646 |
0.12 |
| chr3_57547653_57547804 | 0.21 |
PDE12 |
phosphodiesterase 12 |
5724 |
0.12 |
| chr3_71407403_71407554 | 0.21 |
FOXP1 |
forkhead box P1 |
53567 |
0.15 |
| chr8_129588676_129588964 | 0.21 |
ENSG00000221351 |
. |
243220 |
0.02 |
| chr1_236697521_236697902 | 0.21 |
RP11-385F5.5 |
|
2293 |
0.24 |
| chr2_203533655_203533879 | 0.21 |
FAM117B |
family with sequence similarity 117, member B |
33856 |
0.22 |
| chr2_23509060_23509211 | 0.21 |
KLHL29 |
kelch-like family member 29 |
98953 |
0.09 |
| chr2_12850330_12850505 | 0.21 |
TRIB2 |
tribbles pseudokinase 2 |
6598 |
0.28 |
| chr20_55039592_55039752 | 0.21 |
RTFDC1 |
replication termination factor 2 domain containing 1 |
3975 |
0.15 |
| chr1_180332594_180332745 | 0.21 |
ENSG00000265435 |
. |
74856 |
0.09 |
| chr10_31549661_31549812 | 0.21 |
ENSG00000252479 |
. |
1208 |
0.55 |
| chr12_111889383_111889585 | 0.21 |
ATXN2 |
ataxin 2 |
4432 |
0.21 |
| chr8_22406945_22407789 | 0.20 |
SORBS3 |
sorbin and SH3 domain containing 3 |
1841 |
0.21 |
| chr15_38965016_38965639 | 0.20 |
C15orf53 |
chromosome 15 open reading frame 53 |
23472 |
0.24 |
| chr1_167480391_167480600 | 0.20 |
CD247 |
CD247 molecule |
7280 |
0.21 |
| chr12_107764032_107764308 | 0.20 |
ENSG00000200897 |
. |
4354 |
0.29 |
| chr13_113838414_113838565 | 0.20 |
RP11-98F14.11 |
|
18992 |
0.11 |
| chr1_145432739_145432969 | 0.20 |
TXNIP |
thioredoxin interacting protein |
5615 |
0.13 |
| chr3_11644429_11644663 | 0.20 |
VGLL4 |
vestigial like 4 (Drosophila) |
1509 |
0.4 |
| chrX_99902890_99903180 | 0.20 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
3820 |
0.21 |
| chr1_236579990_236580147 | 0.20 |
EDARADD |
EDAR-associated death domain |
21352 |
0.21 |
| chr2_40325984_40326135 | 0.20 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
5990 |
0.34 |
| chr15_60715225_60715376 | 0.20 |
ANXA2 |
annexin A2 |
20218 |
0.21 |
| chr16_84793876_84794027 | 0.20 |
USP10 |
ubiquitin specific peptidase 10 |
7920 |
0.22 |
| chr2_213980870_213981021 | 0.20 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
32408 |
0.23 |
| chr7_155440870_155441096 | 0.20 |
RBM33 |
RNA binding motif protein 33 |
3587 |
0.25 |
| chr6_106541569_106541720 | 0.20 |
PRDM1 |
PR domain containing 1, with ZNF domain |
4837 |
0.25 |
| chr7_115302535_115302686 | 0.20 |
ENSG00000202377 |
. |
81114 |
0.11 |
| chr7_75111983_75112134 | 0.20 |
POM121C |
POM121 transmembrane nucleoporin C |
1547 |
0.29 |
| chr3_158486550_158486701 | 0.20 |
RP11-379F4.8 |
|
13711 |
0.14 |
| chr2_43793678_43793972 | 0.20 |
THADA |
thyroid adenoma associated |
3829 |
0.29 |
| chr2_42526156_42526376 | 0.20 |
EML4 |
echinoderm microtubule associated protein like 4 |
2127 |
0.4 |
| chrX_46308049_46308200 | 0.20 |
KRBOX4 |
KRAB box domain containing 4 |
456 |
0.88 |
| chr22_40571602_40571800 | 0.20 |
TNRC6B |
trinucleotide repeat containing 6B |
2228 |
0.4 |
| chr5_180243660_180244135 | 0.20 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
1317 |
0.41 |
| chr8_23619073_23619616 | 0.20 |
ENSG00000216123 |
. |
6814 |
0.19 |
| chr2_11480133_11480284 | 0.19 |
ROCK2 |
Rho-associated, coiled-coil containing protein kinase 2 |
4503 |
0.28 |
| chr18_2967412_2967563 | 0.19 |
RP11-737O24.1 |
|
471 |
0.77 |
| chr20_62894793_62894944 | 0.19 |
PCMTD2 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
1997 |
0.4 |
| chr1_33122442_33122593 | 0.19 |
RBBP4 |
retinoblastoma binding protein 4 |
5429 |
0.15 |
| chr2_169090927_169091234 | 0.19 |
STK39 |
serine threonine kinase 39 |
13571 |
0.29 |
| chr16_28567039_28567190 | 0.19 |
CCDC101 |
coiled-coil domain containing 101 |
1867 |
0.24 |
| chr12_66229210_66229361 | 0.19 |
HMGA2 |
high mobility group AT-hook 2 |
10382 |
0.2 |
| chr2_239869911_239870176 | 0.19 |
ENSG00000211566 |
. |
24663 |
0.17 |
| chr5_56099308_56099459 | 0.19 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
12018 |
0.2 |
| chr6_5520923_5521080 | 0.19 |
RP1-232P20.1 |
|
62693 |
0.14 |
| chr15_36654222_36654373 | 0.19 |
C15orf41 |
chromosome 15 open reading frame 41 |
217515 |
0.02 |
| chr19_12400747_12400928 | 0.19 |
ZNF44 |
zinc finger protein 44 |
4791 |
0.18 |
| chrX_152849336_152849542 | 0.19 |
FAM58A |
family with sequence similarity 58, member A |
12025 |
0.13 |
| chr13_74475259_74475410 | 0.19 |
KLF12 |
Kruppel-like factor 12 |
93852 |
0.1 |
| chr10_121028118_121028299 | 0.19 |
ENSG00000242853 |
. |
55839 |
0.09 |
| chr15_52966509_52966798 | 0.19 |
FAM214A |
family with sequence similarity 214, member A |
3765 |
0.26 |
| chr12_66700071_66700537 | 0.19 |
HELB |
helicase (DNA) B |
3979 |
0.2 |
| chr2_102413944_102414095 | 0.19 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
293 |
0.94 |
| chr17_37961333_37961484 | 0.19 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
26930 |
0.12 |
| chr2_179971991_179972202 | 0.19 |
ENSG00000238339 |
. |
18268 |
0.23 |
| chr1_235418994_235419145 | 0.19 |
ARID4B |
AT rich interactive domain 4B (RBP1-like) |
41838 |
0.13 |
| chr11_33290652_33290913 | 0.19 |
HIPK3 |
homeodomain interacting protein kinase 3 |
10905 |
0.22 |
| chr3_20147969_20148120 | 0.19 |
ENSG00000266745 |
. |
31013 |
0.16 |
| chr8_69921640_69921791 | 0.18 |
ENSG00000238808 |
. |
101094 |
0.09 |
| chr10_70751231_70751471 | 0.18 |
KIAA1279 |
KIAA1279 |
2864 |
0.23 |
| chr1_35652154_35652385 | 0.18 |
SFPQ |
splicing factor proline/glutamine-rich |
6480 |
0.19 |
| chr5_55984183_55984556 | 0.18 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
82310 |
0.09 |
| chr16_64230317_64230468 | 0.18 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
863189 |
0.0 |
| chr11_73686038_73686668 | 0.18 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
1698 |
0.28 |
| chr3_45963092_45963243 | 0.18 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
5633 |
0.16 |
| chr2_42404220_42404371 | 0.18 |
AC083949.1 |
|
6854 |
0.23 |
| chr1_193152188_193152339 | 0.18 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
3521 |
0.25 |
| chr3_48004374_48004525 | 0.18 |
ENSG00000199591 |
. |
31045 |
0.16 |
| chr7_130629524_130629906 | 0.18 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
39153 |
0.18 |
| chr17_53847338_53847489 | 0.18 |
PCTP |
phosphatidylcholine transfer protein |
3684 |
0.32 |
| chr13_40894745_40894896 | 0.18 |
ENSG00000252812 |
. |
86757 |
0.09 |
| chr1_31481065_31481216 | 0.18 |
PUM1 |
pumilio RNA-binding family member 1 |
13189 |
0.19 |
| chr2_55757616_55757885 | 0.18 |
CCDC104 |
coiled-coil domain containing 104 |
10883 |
0.15 |
| chr11_93468268_93468502 | 0.18 |
ENSG00000210825 |
. |
17 |
0.89 |
| chr3_114817466_114817617 | 0.18 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
27319 |
0.27 |
| chr14_72231747_72231898 | 0.18 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
146349 |
0.04 |
| chr3_118668683_118668834 | 0.18 |
IGSF11-AS1 |
IGSF11 antisense RNA 1 |
6838 |
0.28 |
| chr3_176900690_176900841 | 0.18 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
13446 |
0.24 |
| chr1_36003036_36003243 | 0.18 |
KIAA0319L |
KIAA0319-like |
17449 |
0.14 |
| chr16_67323885_67324036 | 0.18 |
PLEKHG4 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
2157 |
0.15 |
| chr10_89298476_89298627 | 0.18 |
ENSG00000222192 |
. |
24392 |
0.19 |
| chr21_17656676_17656827 | 0.18 |
ENSG00000201025 |
. |
338 |
0.94 |
| chr3_66330359_66330510 | 0.18 |
ENSG00000206759 |
. |
13210 |
0.22 |
| chr5_77015099_77015250 | 0.18 |
TBCA |
tubulin folding cofactor A |
4032 |
0.32 |
| chr4_186760718_186760869 | 0.18 |
SORBS2 |
sorbin and SH3 domain containing 2 |
26515 |
0.21 |
| chr14_58759101_58759259 | 0.18 |
ENSG00000252782 |
. |
74 |
0.95 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.4 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.0 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:0090266 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.0 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.0 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0002860 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.3 | PID IFNG PATHWAY | IFN-gamma pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.0 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |