Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
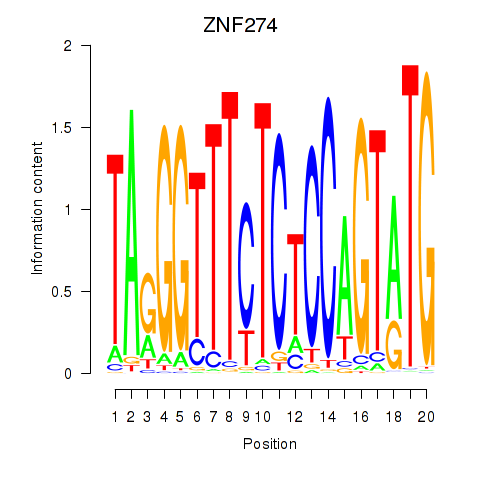
Results for ZNF274
Z-value: 1.31
Transcription factors associated with ZNF274
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF274
|
ENSG00000171606.13 | zinc finger protein 274 |
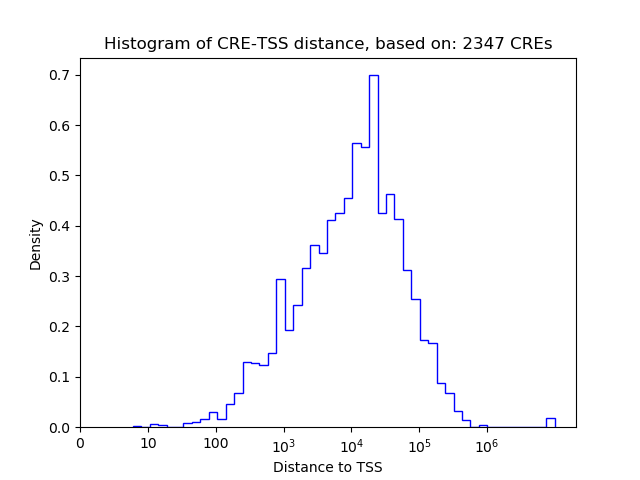
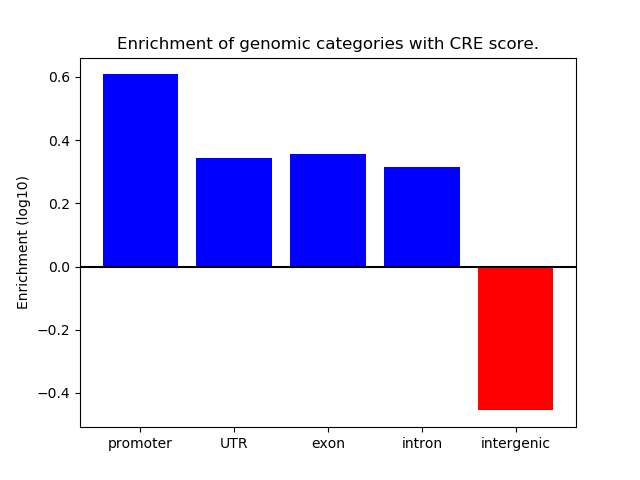
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_58693943_58694152 | ZNF274 | 349 | 0.824205 | -0.56 | 1.2e-01 | Click! |
| chr19_58694466_58694617 | ZNF274 | 145 | 0.938271 | -0.47 | 2.1e-01 | Click! |
| chr19_58695110_58695333 | ZNF274 | 358 | 0.817942 | -0.41 | 2.7e-01 | Click! |
| chr19_58693650_58693919 | ZNF274 | 612 | 0.642528 | -0.33 | 3.9e-01 | Click! |
| chr19_58695365_58695593 | ZNF274 | 616 | 0.639644 | -0.26 | 4.9e-01 | Click! |
Activity of the ZNF274 motif across conditions
Conditions sorted by the z-value of the ZNF274 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
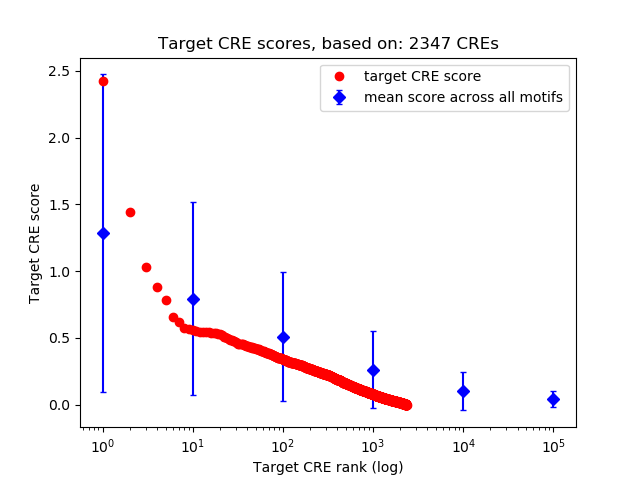
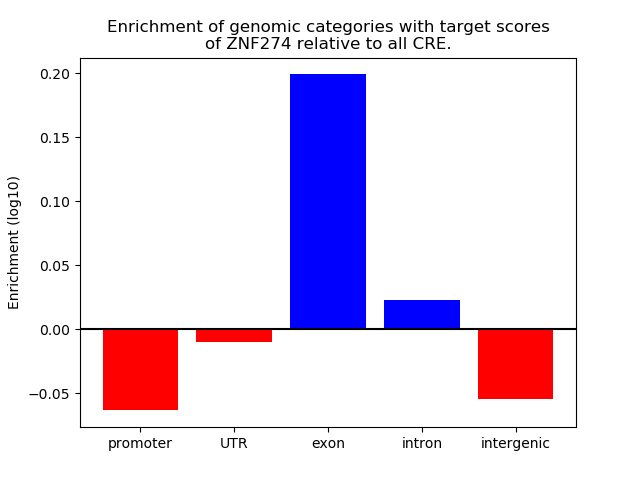
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_89365642_89366074 | 2.42 |
AC137932.5 |
|
1594 |
0.28 |
| chr19_58868146_58868568 | 1.44 |
CTD-2619J13.9 |
|
1008 |
0.25 |
| chr16_89366119_89366411 | 1.03 |
AC137932.5 |
|
2001 |
0.23 |
| chr2_242680975_242681169 | 0.88 |
D2HGDH |
D-2-hydroxyglutarate dehydrogenase |
776 |
0.45 |
| chr7_148767712_148767863 | 0.78 |
ZNF786 |
zinc finger protein 786 |
20010 |
0.12 |
| chr20_24962486_24962637 | 0.66 |
ENSG00000207355 |
. |
7754 |
0.18 |
| chr3_15316866_15317185 | 0.62 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
391 |
0.81 |
| chr17_27068859_27069066 | 0.57 |
AC010761.6 |
|
821 |
0.26 |
| chr7_81396029_81396416 | 0.57 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
3065 |
0.4 |
| chr1_6041214_6041365 | 0.56 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
10237 |
0.18 |
| chr20_418602_419173 | 0.56 |
ENSG00000206797 |
. |
12420 |
0.15 |
| chr11_48037642_48037830 | 0.55 |
AC103828.1 |
|
329 |
0.9 |
| chr6_6726794_6727009 | 0.55 |
LY86-AS1 |
LY86 antisense RNA 1 |
103897 |
0.08 |
| chr7_140058154_140058347 | 0.55 |
SLC37A3 |
solute carrier family 37, member 3 |
6350 |
0.15 |
| chr5_169708109_169708260 | 0.54 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
13853 |
0.22 |
| chr4_88313270_88313528 | 0.54 |
HSD17B11 |
hydroxysteroid (17-beta) dehydrogenase 11 |
861 |
0.54 |
| chr15_90480001_90480190 | 0.54 |
C15orf38 |
chromosome 15 open reading frame 38 |
23907 |
0.1 |
| chr11_122939463_122939751 | 0.53 |
HSPA8 |
heat shock 70kDa protein 8 |
5669 |
0.16 |
| chr20_25655869_25656020 | 0.53 |
ZNF337 |
zinc finger protein 337 |
11402 |
0.16 |
| chr1_10461491_10461642 | 0.53 |
PGD |
phosphogluconate dehydrogenase |
1877 |
0.22 |
| chr3_13050536_13050725 | 0.52 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
22094 |
0.24 |
| chr14_35400530_35400841 | 0.51 |
ENSG00000199980 |
. |
2064 |
0.3 |
| chr13_24852090_24852325 | 0.51 |
SPATA13 |
spermatogenesis associated 13 |
5380 |
0.18 |
| chrX_153214056_153214561 | 0.50 |
RENBP |
renin binding protein |
4076 |
0.1 |
| chr11_10765370_10765523 | 0.49 |
CTR9 |
CTR9, Paf1/RNA polymerase II complex component |
7088 |
0.15 |
| chr13_29156493_29156762 | 0.49 |
POMP |
proteasome maturation protein |
76614 |
0.1 |
| chr16_1538550_1538777 | 0.48 |
PTX4 |
pentraxin 4, long |
195 |
0.86 |
| chr14_91874860_91875123 | 0.48 |
CCDC88C |
coiled-coil domain containing 88C |
8699 |
0.24 |
| chr9_92316642_92316885 | 0.47 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
96810 |
0.09 |
| chr10_104197193_104197463 | 0.47 |
ENSG00000202569 |
. |
1059 |
0.32 |
| chr14_91817459_91817741 | 0.46 |
ENSG00000265856 |
. |
17543 |
0.21 |
| chr5_58568933_58569140 | 0.46 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
2909 |
0.42 |
| chr19_51919928_51920449 | 0.45 |
SIGLEC10 |
sialic acid binding Ig-like lectin 10 |
647 |
0.4 |
| chr13_25200724_25201013 | 0.45 |
ENSG00000211508 |
. |
17651 |
0.18 |
| chr16_87076415_87076642 | 0.45 |
RP11-899L11.3 |
|
172993 |
0.03 |
| chr21_36384069_36384311 | 0.45 |
RUNX1 |
runt-related transcription factor 1 |
37272 |
0.23 |
| chr3_176784016_176784245 | 0.45 |
TBL1XR1-AS1 |
TBL1XR1 antisense RNA 1 |
21481 |
0.22 |
| chr1_25782884_25783035 | 0.45 |
TMEM57 |
transmembrane protein 57 |
25548 |
0.14 |
| chr13_98396488_98396639 | 0.44 |
ENSG00000238407 |
. |
141500 |
0.05 |
| chr12_117033216_117033367 | 0.44 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
19635 |
0.25 |
| chr4_185772755_185773053 | 0.44 |
ENSG00000266698 |
. |
640 |
0.72 |
| chr4_26385783_26386117 | 0.44 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
21798 |
0.26 |
| chrX_37684198_37684679 | 0.44 |
DYNLT3 |
dynein, light chain, Tctex-type 3 |
22223 |
0.19 |
| chr13_111173056_111173211 | 0.44 |
COL4A2-AS1 |
COL4A2 antisense RNA 1 |
12607 |
0.2 |
| chr18_55334626_55334855 | 0.43 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
21160 |
0.15 |
| chr3_71760476_71760839 | 0.43 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
13869 |
0.22 |
| chrX_1590509_1590809 | 0.43 |
ASMTL |
acetylserotonin O-methyltransferase-like |
18004 |
0.18 |
| chrY_1540506_1540807 | 0.42 |
NA |
NA |
> 106 |
NA |
| chr4_124399364_124399515 | 0.42 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
78316 |
0.12 |
| chr1_65472844_65472995 | 0.42 |
ENSG00000212257 |
. |
15838 |
0.19 |
| chr12_11963905_11964056 | 0.42 |
ETV6 |
ets variant 6 |
58545 |
0.15 |
| chr11_67179086_67179395 | 0.42 |
CARNS1 |
carnosine synthase 1 |
3909 |
0.08 |
| chr2_223485669_223485820 | 0.42 |
FARSB |
phenylalanyl-tRNA synthetase, beta subunit |
35026 |
0.15 |
| chr9_140171055_140171601 | 0.42 |
TOR4A |
torsin family 4, member A |
873 |
0.34 |
| chr5_64318013_64318164 | 0.41 |
ENSG00000207439 |
. |
101108 |
0.08 |
| chr1_226985906_226986297 | 0.41 |
ITPKB |
inositol-trisphosphate 3-kinase B |
59077 |
0.12 |
| chr5_131798741_131798941 | 0.41 |
ENSG00000202533 |
. |
4998 |
0.14 |
| chr15_78339587_78339866 | 0.40 |
ENSG00000221476 |
. |
8853 |
0.13 |
| chr21_32561827_32562130 | 0.40 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
59439 |
0.15 |
| chr5_75701847_75702137 | 0.40 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
1743 |
0.49 |
| chr7_105152838_105153078 | 0.40 |
PUS7 |
pseudouridylate synthase 7 homolog (S. cerevisiae) |
4376 |
0.23 |
| chr2_242279172_242279345 | 0.40 |
SEPT2 |
septin 2 |
2367 |
0.22 |
| chr13_94050738_94050889 | 0.40 |
ENSG00000252365 |
. |
29600 |
0.26 |
| chr2_42341709_42341982 | 0.40 |
EML4 |
echinoderm microtubule associated protein like 4 |
54645 |
0.12 |
| chr7_74595648_74595978 | 0.39 |
GTF2IRD2B |
GTF2I repeat domain containing 2B |
38909 |
0.17 |
| chr12_74936306_74936457 | 0.39 |
RP11-56G10.2 |
|
3968 |
0.28 |
| chr2_60673951_60674102 | 0.39 |
AC009970.1 |
|
48795 |
0.14 |
| chr11_121327332_121327753 | 0.39 |
RP11-730K11.1 |
|
3820 |
0.28 |
| chr1_154912617_154913094 | 0.39 |
PMVK |
phosphomevalonate kinase |
3388 |
0.11 |
| chr10_104082217_104082368 | 0.38 |
ENSG00000251989 |
. |
61305 |
0.07 |
| chr10_13386024_13386175 | 0.38 |
SEPHS1 |
selenophosphate synthetase 1 |
3259 |
0.24 |
| chr7_12444012_12444163 | 0.38 |
VWDE |
von Willebrand factor D and EGF domains |
556 |
0.86 |
| chr9_92315939_92316332 | 0.38 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
96182 |
0.09 |
| chr2_175201853_175202044 | 0.38 |
AC018470.1 |
Uncharacterized protein FLJ46347 |
203 |
0.92 |
| chr4_4317352_4317503 | 0.37 |
ZBTB49 |
zinc finger and BTB domain containing 49 |
13073 |
0.18 |
| chr7_30319289_30319440 | 0.37 |
ZNRF2 |
zinc and ring finger 2 |
4559 |
0.23 |
| chr5_157279237_157279388 | 0.37 |
CLINT1 |
clathrin interactor 1 |
6829 |
0.23 |
| chr11_14664037_14664267 | 0.37 |
PSMA1 |
proteasome (prosome, macropain) subunit, alpha type, 1 |
1011 |
0.43 |
| chr3_141194169_141194577 | 0.37 |
RASA2 |
RAS p21 protein activator 2 |
11518 |
0.22 |
| chr2_85895732_85896116 | 0.37 |
SFTPB |
surfactant protein B |
60 |
0.96 |
| chr9_75567554_75567705 | 0.37 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
342 |
0.94 |
| chr18_74212612_74212763 | 0.37 |
RP11-17M16.1 |
uncharacterized protein LOC400658 |
5210 |
0.18 |
| chr21_45303314_45303465 | 0.36 |
AGPAT3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
964 |
0.57 |
| chr10_126228599_126228829 | 0.36 |
LHPP |
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
78302 |
0.09 |
| chr2_114321807_114321958 | 0.36 |
ENSG00000221055 |
. |
18791 |
0.16 |
| chr1_17956374_17956572 | 0.36 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
11624 |
0.28 |
| chr12_21831996_21832380 | 0.36 |
RP11-59N23.3 |
|
16941 |
0.18 |
| chr12_75851152_75851303 | 0.35 |
GLIPR1 |
GLI pathogenesis-related 1 |
23233 |
0.16 |
| chr5_171427537_171427688 | 0.35 |
FBXW11 |
F-box and WD repeat domain containing 11 |
3621 |
0.3 |
| chr4_90878223_90878385 | 0.35 |
MMRN1 |
multimerin 1 |
55170 |
0.16 |
| chr17_14220339_14220600 | 0.35 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
16069 |
0.26 |
| chr14_22952984_22953135 | 0.35 |
ENSG00000251002 |
. |
1111 |
0.3 |
| chr7_36325933_36326467 | 0.35 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
10553 |
0.18 |
| chr17_19273582_19274475 | 0.35 |
MAPK7 |
mitogen-activated protein kinase 7 |
7006 |
0.11 |
| chr21_36410531_36410893 | 0.35 |
RUNX1 |
runt-related transcription factor 1 |
10750 |
0.32 |
| chr12_92478066_92478217 | 0.35 |
C12orf79 |
chromosome 12 open reading frame 79 |
52656 |
0.13 |
| chrX_119737541_119737692 | 0.35 |
MCTS1 |
malignant T cell amplified sequence 1 |
219 |
0.93 |
| chr10_90747212_90747556 | 0.34 |
FAS |
Fas cell surface death receptor |
3030 |
0.19 |
| chr22_24830948_24831363 | 0.34 |
ADORA2A |
adenosine A2a receptor |
2347 |
0.28 |
| chr8_102713284_102713482 | 0.34 |
KB-1107E3.1 |
|
14608 |
0.22 |
| chr1_203294006_203294279 | 0.34 |
ENSG00000202300 |
. |
5915 |
0.19 |
| chr7_13133751_13133902 | 0.34 |
ENSG00000222974 |
. |
217517 |
0.02 |
| chr12_32419800_32419951 | 0.34 |
BICD1 |
bicaudal D homolog 1 (Drosophila) |
61461 |
0.13 |
| chr3_4559229_4559481 | 0.34 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
1179 |
0.55 |
| chr2_135031193_135031344 | 0.33 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
19438 |
0.24 |
| chr11_113997404_113997555 | 0.33 |
ENSG00000221112 |
. |
39828 |
0.17 |
| chr16_74560683_74560834 | 0.33 |
GLG1 |
golgi glycoprotein 1 |
36465 |
0.15 |
| chr7_38714149_38714300 | 0.33 |
FAM183B |
family with sequence similarity 183, member B |
12413 |
0.24 |
| chr3_47099072_47099238 | 0.33 |
ENSG00000251938 |
. |
30943 |
0.15 |
| chr2_42523506_42523974 | 0.33 |
EML4 |
echinoderm microtubule associated protein like 4 |
4653 |
0.29 |
| chr1_181059536_181059906 | 0.33 |
IER5 |
immediate early response 5 |
2083 |
0.37 |
| chr13_51185358_51185546 | 0.33 |
DLEU7-AS1 |
DLEU7 antisense RNA 1 |
196540 |
0.03 |
| chr12_105478171_105478322 | 0.32 |
ALDH1L2 |
aldehyde dehydrogenase 1 family, member L2 |
95 |
0.96 |
| chr7_87943895_87944046 | 0.32 |
STEAP4 |
STEAP family member 4 |
7764 |
0.24 |
| chr2_3083933_3084084 | 0.32 |
ENSG00000238722 |
. |
4464 |
0.32 |
| chr10_90686436_90686600 | 0.32 |
ACTA2-AS1 |
ACTA2 antisense RNA 1 |
5882 |
0.17 |
| chr21_19186267_19186567 | 0.32 |
C21orf91 |
chromosome 21 open reading frame 91 |
4919 |
0.23 |
| chr4_24980289_24980589 | 0.32 |
CCDC149 |
coiled-coil domain containing 149 |
1331 |
0.55 |
| chr20_40008439_40008590 | 0.32 |
EMILIN3 |
elastin microfibril interfacer 3 |
13047 |
0.2 |
| chr2_158290506_158290794 | 0.32 |
CYTIP |
cytohesin 1 interacting protein |
5276 |
0.24 |
| chr19_15625593_15626001 | 0.32 |
CYP4F22 |
cytochrome P450, family 4, subfamily F, polypeptide 22 |
6493 |
0.15 |
| chr2_120285078_120285229 | 0.32 |
SCTR |
secretin receptor |
3083 |
0.24 |
| chr18_33199033_33199184 | 0.32 |
RP11-712P20.2 |
|
23243 |
0.2 |
| chr7_148519000_148519151 | 0.32 |
ENSG00000251712 |
. |
971 |
0.58 |
| chr6_119674802_119674953 | 0.32 |
MAN1A1 |
mannosidase, alpha, class 1A, member 1 |
3951 |
0.3 |
| chr9_36145819_36145970 | 0.32 |
GLIPR2 |
GLI pathogenesis-related 2 |
9152 |
0.18 |
| chr2_88210407_88210558 | 0.31 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
74827 |
0.09 |
| chr12_102276162_102276313 | 0.31 |
DRAM1 |
DNA-damage regulated autophagy modulator 1 |
4776 |
0.16 |
| chr9_117126749_117127269 | 0.31 |
AKNA |
AT-hook transcription factor |
12235 |
0.19 |
| chr4_9428848_9428999 | 0.31 |
ENSG00000252307 |
. |
15944 |
0.11 |
| chr2_68385602_68386003 | 0.31 |
PNO1 |
partner of NOB1 homolog (S. cerevisiae) |
826 |
0.45 |
| chr9_51365_51516 | 0.31 |
RP11-143M1.2 |
|
21261 |
0.19 |
| chr11_48065545_48066080 | 0.31 |
AC103828.1 |
|
28405 |
0.17 |
| chr9_4325119_4325384 | 0.31 |
GLIS3 |
GLIS family zinc finger 3 |
25335 |
0.17 |
| chr2_144235573_144235784 | 0.31 |
AC096558.1 |
|
2672 |
0.38 |
| chr10_134403005_134403189 | 0.31 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
18333 |
0.23 |
| chr9_79234557_79234812 | 0.31 |
PRUNE2 |
prune homolog 2 (Drosophila) |
32781 |
0.17 |
| chr18_32624943_32625387 | 0.31 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
3551 |
0.36 |
| chr2_84684670_84684900 | 0.31 |
SUCLG1 |
succinate-CoA ligase, alpha subunit |
1819 |
0.48 |
| chr13_47992322_47992473 | 0.31 |
ENSG00000244521 |
. |
40972 |
0.23 |
| chr2_237886238_237886389 | 0.31 |
ENSG00000202341 |
. |
37635 |
0.18 |
| chr20_37184274_37184425 | 0.30 |
ADIG |
adipogenin |
25489 |
0.13 |
| chr3_113955237_113955388 | 0.30 |
ZNF80 |
zinc finger protein 80 |
1113 |
0.48 |
| chr4_175029310_175029461 | 0.30 |
FBXO8 |
F-box protein 8 |
175429 |
0.03 |
| chr9_5846551_5846725 | 0.30 |
ERMP1 |
endoplasmic reticulum metallopeptidase 1 |
13521 |
0.19 |
| chr1_55660397_55660593 | 0.30 |
USP24 |
ubiquitin specific peptidase 24 |
20267 |
0.21 |
| chr2_7197696_7198047 | 0.30 |
AC019048.1 |
|
20140 |
0.23 |
| chr10_65012309_65012460 | 0.30 |
JMJD1C |
jumonji domain containing 1C |
16442 |
0.26 |
| chr15_63804900_63805051 | 0.30 |
USP3 |
ubiquitin specific peptidase 3 |
2670 |
0.32 |
| chr3_3213817_3214181 | 0.30 |
CRBN |
cereblon |
7359 |
0.19 |
| chr13_74289274_74289507 | 0.30 |
KLF12 |
Kruppel-like factor 12 |
279796 |
0.01 |
| chr2_54004004_54004155 | 0.30 |
CHAC2 |
ChaC, cation transport regulator homolog 2 (E. coli) |
9150 |
0.15 |
| chr5_57787468_57787619 | 0.30 |
GAPT |
GRB2-binding adaptor protein, transmembrane |
279 |
0.92 |
| chr6_111134072_111134223 | 0.30 |
CDK19 |
cyclin-dependent kinase 19 |
2152 |
0.3 |
| chr20_30653904_30654055 | 0.30 |
HCK |
hemopoietic cell kinase |
13915 |
0.12 |
| chr14_75612464_75612670 | 0.30 |
NEK9 |
NIMA-related kinase 9 |
18520 |
0.13 |
| chr6_112046537_112046688 | 0.30 |
FYN |
FYN oncogene related to SRC, FGR, YES |
5347 |
0.28 |
| chr8_126521344_126521495 | 0.30 |
ENSG00000266452 |
. |
64612 |
0.13 |
| chr4_73257014_73257165 | 0.30 |
RP11-373J21.1 |
|
68344 |
0.14 |
| chr3_148898157_148898308 | 0.30 |
CP |
ceruloplasmin (ferroxidase) |
824 |
0.65 |
| chr21_46530039_46530452 | 0.29 |
PRED58 |
|
4628 |
0.17 |
| chr11_5818563_5818714 | 0.29 |
OR52N1 |
olfactory receptor, family 52, subfamily N, member 1 |
8592 |
0.12 |
| chr7_128720941_128721092 | 0.29 |
ENSG00000238733 |
. |
19431 |
0.15 |
| chr9_4766664_4766815 | 0.29 |
AK3 |
adenylate kinase 3 |
24696 |
0.15 |
| chr2_225799627_225799778 | 0.29 |
DOCK10 |
dedicator of cytokinesis 10 |
12080 |
0.29 |
| chr15_74224861_74225012 | 0.29 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
4347 |
0.17 |
| chr9_112920768_112920919 | 0.29 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
33062 |
0.19 |
| chr5_126907475_126907626 | 0.29 |
PRRC1 |
proline-rich coiled-coil 1 |
54202 |
0.15 |
| chr16_31469602_31469809 | 0.29 |
ARMC5 |
armadillo repeat containing 5 |
304 |
0.75 |
| chr8_145239667_145240434 | 0.29 |
MROH1 |
maestro heat-like repeat family member 1 |
21377 |
0.11 |
| chr7_130780107_130780479 | 0.29 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
7060 |
0.22 |
| chr9_134146913_134147198 | 0.29 |
FAM78A |
family with sequence similarity 78, member A |
1145 |
0.48 |
| chr1_23852013_23852546 | 0.29 |
E2F2 |
E2F transcription factor 2 |
5433 |
0.19 |
| chr16_31075251_31075402 | 0.28 |
ZNF668 |
zinc finger protein 668 |
964 |
0.26 |
| chr13_100635255_100635469 | 0.28 |
ZIC2 |
Zic family member 2 |
1336 |
0.43 |
| chr16_12175053_12175238 | 0.28 |
RP11-276H1.3 |
|
9014 |
0.17 |
| chr2_114457524_114457975 | 0.28 |
ENSG00000202427 |
. |
19061 |
0.16 |
| chr3_108047727_108047992 | 0.28 |
ENSG00000221724 |
. |
8620 |
0.23 |
| chr8_135033119_135033449 | 0.28 |
ENSG00000212273 |
. |
376316 |
0.01 |
| chr1_19455827_19455978 | 0.28 |
UBR4 |
ubiquitin protein ligase E3 component n-recognin 4 |
10559 |
0.2 |
| chr15_53847423_53847574 | 0.28 |
ENSG00000252066 |
. |
96676 |
0.09 |
| chr21_47893746_47893897 | 0.28 |
DIP2A-IT1 |
DIP2A intronic transcript 1 (non-protein coding) |
11437 |
0.16 |
| chr5_157251804_157251955 | 0.28 |
ENSG00000206819 |
. |
15773 |
0.18 |
| chrX_18513070_18513221 | 0.28 |
CDKL5 |
cyclin-dependent kinase-like 5 |
52837 |
0.15 |
| chr1_92954823_92954974 | 0.28 |
GFI1 |
growth factor independent 1 transcription repressor |
2465 |
0.38 |
| chr4_153030332_153030483 | 0.28 |
ENSG00000266244 |
. |
119213 |
0.06 |
| chr17_17664043_17664194 | 0.28 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
10017 |
0.13 |
| chr6_24962830_24963080 | 0.28 |
FAM65B |
family with sequence similarity 65, member B |
26767 |
0.19 |
| chr8_9450617_9450846 | 0.28 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
18214 |
0.23 |
| chrX_109351476_109351643 | 0.27 |
ENSG00000265584 |
. |
26213 |
0.2 |
| chr21_36416402_36416644 | 0.27 |
RUNX1 |
runt-related transcription factor 1 |
4939 |
0.36 |
| chr5_138720278_138720769 | 0.27 |
SLC23A1 |
solute carrier family 23 (ascorbic acid transporter), member 1 |
281 |
0.82 |
| chr18_76930724_76930875 | 0.27 |
CTD-2286N8.2 |
|
53592 |
0.15 |
| chr4_48265932_48266083 | 0.27 |
TEC |
tec protein tyrosine kinase |
5874 |
0.26 |
| chr10_46934055_46934235 | 0.27 |
RP11-38L15.3 |
|
17327 |
0.15 |
| chr4_177557559_177557710 | 0.27 |
RP11-313E19.2 |
|
33138 |
0.24 |
| chr20_43782810_43782961 | 0.27 |
PI3 |
peptidase inhibitor 3, skin-derived |
20632 |
0.12 |
| chr11_128355434_128355619 | 0.27 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
19763 |
0.23 |
| chr3_169498435_169498680 | 0.27 |
RP11-362K14.5 |
|
435 |
0.67 |
| chr6_34985267_34985418 | 0.27 |
TCP11 |
t-complex 11, testis-specific |
103480 |
0.06 |
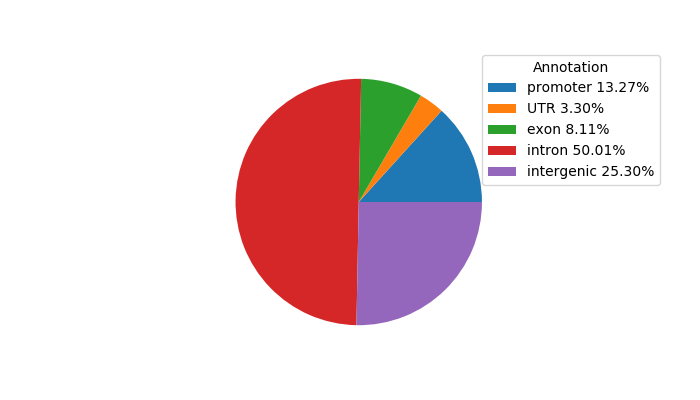
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 0.4 | GO:1902170 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.1 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.2 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.3 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.1 | 0.3 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.1 | 0.2 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.2 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.1 | 0.2 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.2 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.4 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.2 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.2 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:2000351 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.2 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0043301 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.0 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.0 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.0 | GO:0052031 | modulation by symbiont of host defense response(GO:0052031) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0046348 | amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.0 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.4 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.5 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.0 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.3 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.0 | 0.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.2 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.0 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.7 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.2 | GO:0043022 | ribosome binding(GO:0043022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.0 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |