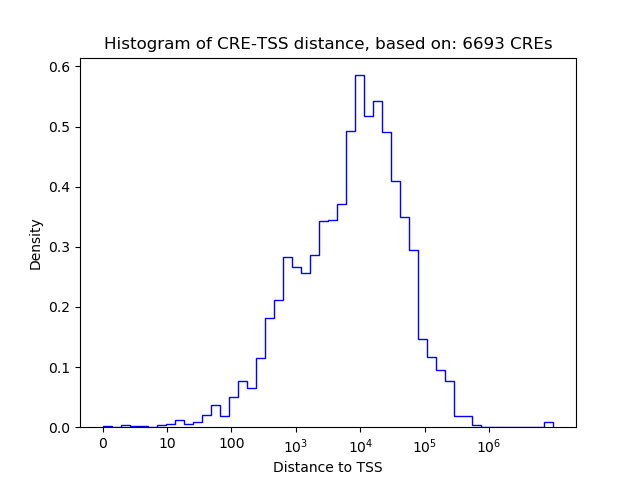
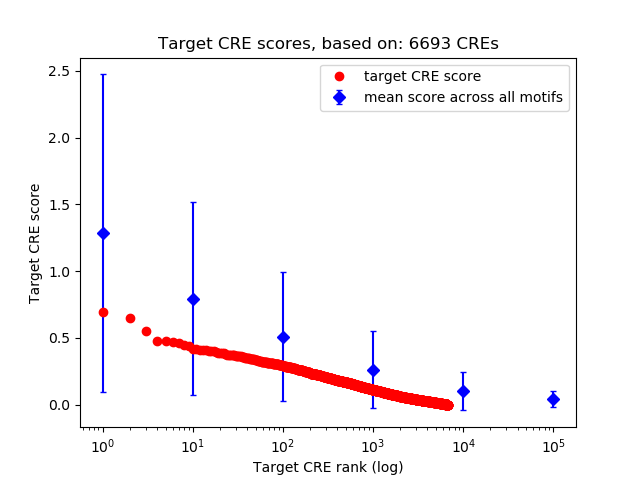
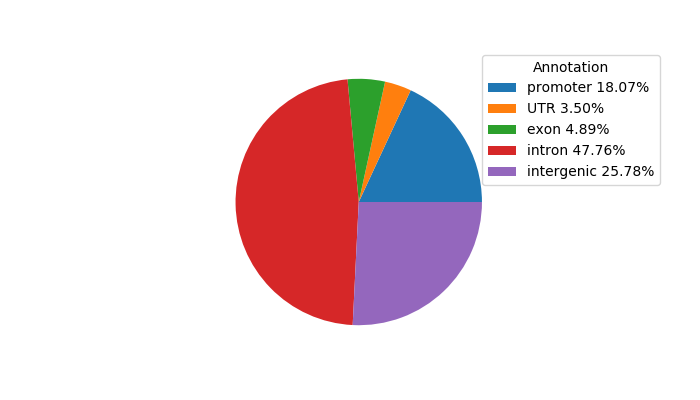
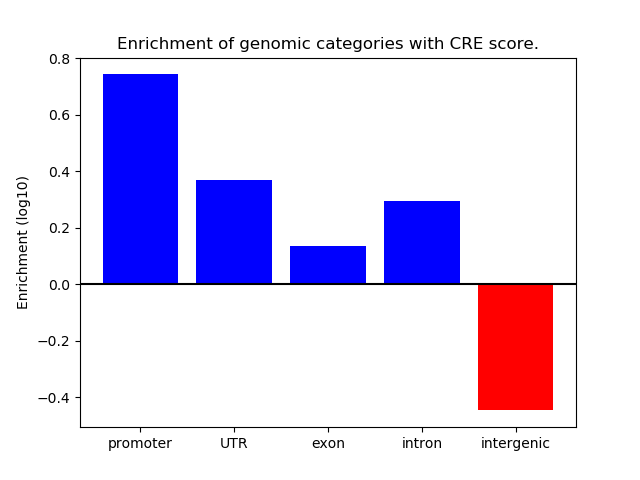
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for ZNF282
Z-value: 1.24
Transcription factors associated with ZNF282
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF282
|
ENSG00000170265.7 | zinc finger protein 282 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_148893289_148893440 | ZNF282 | 722 | 0.627642 | 0.90 | 9.7e-04 | Click! |
| chr7_148893510_148893675 | ZNF282 | 950 | 0.516032 | 0.87 | 2.5e-03 | Click! |
| chr7_148878722_148878873 | ZNF282 | 13780 | 0.144296 | -0.74 | 2.1e-02 | Click! |
| chr7_148888859_148889047 | ZNF282 | 3624 | 0.195190 | 0.69 | 3.9e-02 | Click! |
| chr7_148877933_148878084 | ZNF282 | 14569 | 0.142631 | -0.67 | 4.9e-02 | Click! |
Activity of the ZNF282 motif across conditions
Conditions sorted by the z-value of the ZNF282 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_46572017_46572193 | 0.70 |
SLC38A1 |
solute carrier family 38, member 1 |
89379 |
0.1 |
| chr2_96830073_96830240 | 0.65 |
DUSP2 |
dual specificity phosphatase 2 |
18977 |
0.13 |
| chr7_142502062_142502213 | 0.55 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
21006 |
0.16 |
| chr10_82288993_82289258 | 0.48 |
RP11-137H2.4 |
|
6573 |
0.22 |
| chr1_109188369_109188646 | 0.47 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
15170 |
0.19 |
| chr12_6573393_6573619 | 0.47 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
6305 |
0.09 |
| chr20_13619077_13619233 | 0.46 |
TASP1 |
taspase, threonine aspartase, 1 |
395 |
0.92 |
| chr6_36067796_36067952 | 0.45 |
MAPK13 |
mitogen-activated protein kinase 13 |
30224 |
0.14 |
| chr7_3020531_3020974 | 0.44 |
ENSG00000201794 |
. |
17976 |
0.2 |
| chr2_55757616_55757885 | 0.42 |
CCDC104 |
coiled-coil domain containing 104 |
10883 |
0.15 |
| chr11_14292192_14292493 | 0.42 |
RP11-21L19.1 |
|
2895 |
0.33 |
| chr2_234308315_234308480 | 0.41 |
DGKD |
diacylglycerol kinase, delta 130kDa |
11597 |
0.17 |
| chr14_91302849_91303150 | 0.41 |
TTC7B |
tetratricopeptide repeat domain 7B |
20176 |
0.25 |
| chr13_30944650_30944977 | 0.41 |
KATNAL1 |
katanin p60 subunit A-like 1 |
63192 |
0.13 |
| chr2_7018212_7018651 | 0.40 |
RSAD2 |
radical S-adenosyl methionine domain containing 2 |
494 |
0.78 |
| chr8_82044607_82044805 | 0.40 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
20403 |
0.26 |
| chr15_91143512_91143728 | 0.40 |
CTD-3065B20.2 |
|
3941 |
0.18 |
| chr10_7237970_7238148 | 0.40 |
SFMBT2 |
Scm-like with four mbt domains 2 |
212648 |
0.02 |
| chr1_117310626_117310883 | 0.39 |
CD2 |
CD2 molecule |
13665 |
0.21 |
| chrX_9286819_9287136 | 0.39 |
TBL1X |
transducin (beta)-like 1X-linked |
144358 |
0.05 |
| chr18_60880123_60880495 | 0.39 |
ENSG00000238988 |
. |
18411 |
0.21 |
| chr19_47948776_47948927 | 0.39 |
MEIS3 |
Meis homeobox 3 |
26071 |
0.11 |
| chr14_89817017_89817302 | 0.38 |
RP11-356K23.1 |
|
513 |
0.77 |
| chr1_111433483_111434153 | 0.37 |
CD53 |
CD53 molecule |
18042 |
0.16 |
| chrX_103367091_103367582 | 0.37 |
ZCCHC18 |
zinc finger, CCHC domain containing 18 |
10134 |
0.18 |
| chr20_50097371_50097649 | 0.37 |
ENSG00000266761 |
. |
27996 |
0.21 |
| chr7_150200817_150201275 | 0.37 |
GIMAP7 |
GTPase, IMAP family member 7 |
10872 |
0.18 |
| chr17_37252182_37252943 | 0.37 |
PLXDC1 |
plexin domain containing 1 |
11178 |
0.15 |
| chr7_142378873_142379024 | 0.37 |
MTRNR2L6 |
MT-RNR2-like 6 |
4844 |
0.28 |
| chr8_129672236_129672673 | 0.37 |
ENSG00000221351 |
. |
159586 |
0.04 |
| chr1_101745654_101745926 | 0.37 |
RP4-575N6.5 |
|
37076 |
0.14 |
| chr1_117903396_117903547 | 0.37 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
6600 |
0.31 |
| chr9_20373431_20373643 | 0.36 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
8930 |
0.21 |
| chr16_84119947_84120139 | 0.36 |
MBTPS1 |
membrane-bound transcription factor peptidase, site 1 |
30357 |
0.1 |
| chr12_65672789_65672940 | 0.36 |
MSRB3 |
methionine sulfoxide reductase B3 |
97 |
0.97 |
| chr3_50611684_50611927 | 0.36 |
HEMK1 |
HemK methyltransferase family member 1 |
2750 |
0.2 |
| chr17_37911699_37911850 | 0.36 |
GRB7 |
growth factor receptor-bound protein 7 |
13269 |
0.11 |
| chr4_2787896_2788215 | 0.35 |
SH3BP2 |
SH3-domain binding protein 2 |
6695 |
0.21 |
| chr18_2636269_2636739 | 0.35 |
ENSG00000200875 |
. |
13362 |
0.14 |
| chr3_32411752_32412018 | 0.35 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
21278 |
0.22 |
| chr5_156630874_156631025 | 0.35 |
CTB-4E7.1 |
|
20441 |
0.11 |
| chr4_108984413_108984564 | 0.35 |
HADH |
hydroxyacyl-CoA dehydrogenase |
58697 |
0.12 |
| chr6_37141344_37141568 | 0.35 |
PIM1 |
pim-1 oncogene |
3477 |
0.23 |
| chrX_135839770_135839921 | 0.34 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
9657 |
0.18 |
| chr5_158239944_158240095 | 0.34 |
CTD-2363C16.1 |
|
169995 |
0.04 |
| chr15_38982599_38982925 | 0.34 |
C15orf53 |
chromosome 15 open reading frame 53 |
6037 |
0.31 |
| chr17_17660810_17661016 | 0.34 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
13222 |
0.13 |
| chr6_36005080_36005248 | 0.34 |
MAPK14 |
mitogen-activated protein kinase 14 |
8298 |
0.19 |
| chr18_29606522_29606811 | 0.34 |
RNF125 |
ring finger protein 125, E3 ubiquitin protein ligase |
8331 |
0.16 |
| chr5_67577719_67577949 | 0.34 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
1699 |
0.51 |
| chr9_116288167_116288318 | 0.33 |
RGS3 |
regulator of G-protein signaling 3 |
8692 |
0.23 |
| chr1_245020022_245020173 | 0.33 |
HNRNPU-AS1 |
HNRNPU antisense RNA 1 |
1298 |
0.41 |
| chr3_13462874_13463150 | 0.33 |
NUP210 |
nucleoporin 210kDa |
1203 |
0.55 |
| chr3_4334985_4335136 | 0.33 |
SETMAR |
SET domain and mariner transposase fusion gene |
9928 |
0.31 |
| chr14_103368353_103368504 | 0.33 |
AMN |
amnion associated transmembrane protein |
20565 |
0.12 |
| chr3_13048597_13048870 | 0.33 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
20197 |
0.25 |
| chr3_38005322_38005473 | 0.32 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
673 |
0.64 |
| chr16_80598344_80598524 | 0.32 |
RP11-525K10.3 |
|
1402 |
0.4 |
| chr1_235291589_235291740 | 0.32 |
ENSG00000207181 |
. |
412 |
0.58 |
| chr1_54946775_54946928 | 0.32 |
ENSG00000265404 |
. |
23490 |
0.17 |
| chr12_123592299_123592450 | 0.32 |
PITPNM2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
2601 |
0.28 |
| chr5_14794238_14794389 | 0.32 |
ENSG00000264792 |
. |
31808 |
0.19 |
| chr15_52046369_52046544 | 0.32 |
CTD-2308G16.1 |
|
1774 |
0.24 |
| chr3_15308861_15309012 | 0.32 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
8480 |
0.14 |
| chrY_2874457_2874608 | 0.32 |
ZFY-AS1 |
ZFY antisense RNA 1 |
3865 |
0.34 |
| chr5_169704104_169704255 | 0.32 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
9848 |
0.22 |
| chr3_186678062_186678218 | 0.32 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
14605 |
0.21 |
| chr1_27944687_27944855 | 0.32 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
5802 |
0.15 |
| chr22_22603206_22603357 | 0.31 |
VPREB1 |
pre-B lymphocyte 1 |
4089 |
0.1 |
| chr5_14701641_14701810 | 0.31 |
FAM105B |
family with sequence similarity 105, member B |
20066 |
0.2 |
| chr2_95688672_95688933 | 0.31 |
MAL |
mal, T-cell differentiation protein |
2627 |
0.21 |
| chr17_35447597_35447771 | 0.31 |
ACACA |
acetyl-CoA carboxylase alpha |
1766 |
0.34 |
| chr14_105532821_105533069 | 0.31 |
GPR132 |
G protein-coupled receptor 132 |
1163 |
0.49 |
| chr11_124813257_124813486 | 0.31 |
HEPACAM |
hepatic and glial cell adhesion molecule |
7063 |
0.13 |
| chr5_39175704_39176163 | 0.31 |
FYB |
FYN binding protein |
27196 |
0.22 |
| chr17_14221651_14221911 | 0.31 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
17381 |
0.25 |
| chr11_118182213_118182479 | 0.31 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
6732 |
0.13 |
| chr13_42070374_42070546 | 0.31 |
RGCC |
regulator of cell cycle |
38765 |
0.13 |
| chr3_43343792_43343998 | 0.31 |
ENSG00000241939 |
. |
12548 |
0.17 |
| chr18_13465523_13465878 | 0.31 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
686 |
0.52 |
| chr21_36409118_36409316 | 0.31 |
RUNX1 |
runt-related transcription factor 1 |
12245 |
0.31 |
| chr19_6775713_6775940 | 0.30 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
2857 |
0.17 |
| chr2_169290019_169290449 | 0.30 |
ENSG00000239230 |
. |
7481 |
0.21 |
| chr13_22032610_22032761 | 0.30 |
ZDHHC20 |
zinc finger, DHHC-type containing 20 |
729 |
0.72 |
| chr12_11697430_11697755 | 0.30 |
ENSG00000251747 |
. |
1821 |
0.33 |
| chr10_98516773_98516924 | 0.30 |
ENSG00000207287 |
. |
13924 |
0.19 |
| chr7_105525970_105526305 | 0.30 |
ATXN7L1 |
ataxin 7-like 1 |
9087 |
0.29 |
| chr8_21772448_21772871 | 0.30 |
DOK2 |
docking protein 2, 56kDa |
1288 |
0.44 |
| chr21_43658442_43658806 | 0.30 |
ENSG00000223262 |
. |
17341 |
0.16 |
| chr3_18412272_18412602 | 0.30 |
RP11-158G18.1 |
|
38690 |
0.18 |
| chr4_40230061_40230375 | 0.30 |
RHOH |
ras homolog family member H |
28254 |
0.18 |
| chr2_241342781_241342932 | 0.30 |
GPC1 |
glypican 1 |
32232 |
0.15 |
| chr15_64128117_64128268 | 0.30 |
HERC1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
2051 |
0.37 |
| chr2_8596031_8596222 | 0.30 |
AC011747.7 |
|
219770 |
0.02 |
| chr1_12405618_12405821 | 0.30 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
64205 |
0.11 |
| chr19_55764964_55765151 | 0.30 |
PPP6R1 |
protein phosphatase 6, regulatory subunit 1 |
2080 |
0.15 |
| chr1_121347938_121348091 | 0.30 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
412077 |
0.01 |
| chr20_25676704_25676934 | 0.29 |
ZNF337 |
zinc finger protein 337 |
658 |
0.71 |
| chr2_198075189_198075340 | 0.29 |
ANKRD44 |
ankyrin repeat domain 44 |
12502 |
0.2 |
| chr7_138728882_138729132 | 0.29 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
8232 |
0.21 |
| chr12_104912220_104912483 | 0.29 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
61572 |
0.13 |
| chr6_41976653_41976895 | 0.29 |
ENSG00000206875 |
. |
9478 |
0.16 |
| chr19_19051022_19051173 | 0.29 |
HOMER3 |
homer homolog 3 (Drosophila) |
16 |
0.5 |
| chr5_130852523_130852701 | 0.29 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
8762 |
0.3 |
| chr18_21059346_21059497 | 0.29 |
RIOK3 |
RIO kinase 3 |
2269 |
0.26 |
| chr11_121416006_121416228 | 0.29 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
21317 |
0.27 |
| chr10_11364003_11364154 | 0.29 |
CELF2-AS1 |
CELF2 antisense RNA 1 |
2231 |
0.44 |
| chr9_134145646_134145833 | 0.29 |
FAM78A |
family with sequence similarity 78, member A |
141 |
0.96 |
| chr9_37075041_37075192 | 0.29 |
RP11-220I1.2 |
|
40371 |
0.13 |
| chr10_112603695_112603846 | 0.29 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
27795 |
0.13 |
| chr6_37469689_37469840 | 0.28 |
CCDC167 |
coiled-coil domain containing 167 |
2066 |
0.34 |
| chr2_143975242_143975418 | 0.28 |
RP11-190J23.1 |
|
45589 |
0.18 |
| chr17_78721062_78721213 | 0.28 |
RP11-28G8.1 |
|
58295 |
0.13 |
| chr13_52394867_52395060 | 0.28 |
RP11-327P2.5 |
|
16530 |
0.18 |
| chr8_91640248_91640450 | 0.28 |
TMEM64 |
transmembrane protein 64 |
17391 |
0.21 |
| chr12_54675160_54675465 | 0.28 |
HNRNPA1 |
heterogeneous nuclear ribonucleoprotein A1 |
342 |
0.7 |
| chr2_225895814_225896091 | 0.28 |
DOCK10 |
dedicator of cytokinesis 10 |
11207 |
0.25 |
| chr2_106380015_106380293 | 0.28 |
NCK2 |
NCK adaptor protein 2 |
17966 |
0.27 |
| chr18_46586685_46586836 | 0.28 |
ENSG00000263849 |
. |
10622 |
0.19 |
| chr5_54398513_54398753 | 0.28 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
157 |
0.94 |
| chr4_84028872_84029038 | 0.28 |
PLAC8 |
placenta-specific 8 |
2041 |
0.38 |
| chr1_208004097_208004248 | 0.28 |
ENSG00000203709 |
. |
28304 |
0.2 |
| chr12_7344185_7344336 | 0.28 |
PEX5 |
peroxisomal biogenesis factor 5 |
852 |
0.49 |
| chr8_61812488_61812744 | 0.28 |
RP11-33I11.2 |
|
90451 |
0.09 |
| chr20_31000245_31000406 | 0.28 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
18340 |
0.18 |
| chr9_77767149_77767402 | 0.28 |
ENSG00000200041 |
. |
29262 |
0.16 |
| chr6_166832217_166832454 | 0.28 |
RP1-168L15.5 |
|
35658 |
0.13 |
| chr10_11236479_11236664 | 0.28 |
RP3-323N1.2 |
|
23232 |
0.2 |
| chr5_133466460_133466611 | 0.28 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
7226 |
0.22 |
| chrX_20264408_20264559 | 0.28 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
4677 |
0.28 |
| chr1_246984990_246985280 | 0.27 |
ENSG00000252011 |
. |
11696 |
0.16 |
| chr1_160832817_160833049 | 0.27 |
CD244 |
CD244 molecule, natural killer cell receptor 2B4 |
241 |
0.91 |
| chr2_74248551_74248821 | 0.27 |
TET3 |
tet methylcytosine dioxygenase 3 |
18846 |
0.16 |
| chr9_96930007_96930263 | 0.27 |
ENSG00000199165 |
. |
8099 |
0.16 |
| chr15_45879124_45879275 | 0.27 |
BLOC1S6 |
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
122 |
0.76 |
| chr11_14269963_14270251 | 0.27 |
RP11-21L19.1 |
|
25130 |
0.22 |
| chr14_99672150_99672346 | 0.27 |
AL162151.4 |
|
47495 |
0.14 |
| chr12_102101528_102101679 | 0.27 |
CHPT1 |
choline phosphotransferase 1 |
10153 |
0.13 |
| chr22_40571822_40572044 | 0.27 |
TNRC6B |
trinucleotide repeat containing 6B |
1996 |
0.43 |
| chr6_47276688_47276935 | 0.27 |
TNFRSF21 |
tumor necrosis factor receptor superfamily, member 21 |
830 |
0.75 |
| chr21_46271927_46272078 | 0.27 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
9846 |
0.13 |
| chr21_45509719_45509870 | 0.27 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
17377 |
0.16 |
| chr5_175079178_175079630 | 0.27 |
HRH2 |
histamine receptor H2 |
5629 |
0.23 |
| chr5_133426970_133427238 | 0.26 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
23298 |
0.2 |
| chr10_30783764_30783915 | 0.26 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
56088 |
0.12 |
| chr11_64560688_64561099 | 0.26 |
MAP4K2 |
mitogen-activated protein kinase kinase kinase kinase 2 |
9754 |
0.11 |
| chr17_38760477_38760676 | 0.26 |
SMARCE1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
27966 |
0.12 |
| chr1_162468126_162468277 | 0.26 |
UHMK1 |
U2AF homology motif (UHM) kinase 1 |
568 |
0.76 |
| chr15_60864333_60864586 | 0.26 |
RORA |
RAR-related orphan receptor A |
20281 |
0.19 |
| chr1_27118119_27118479 | 0.26 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
2880 |
0.17 |
| chr11_67368212_67368440 | 0.26 |
RP11-655M14.12 |
|
4666 |
0.1 |
| chr17_26940722_26940873 | 0.26 |
SGK494 |
uncharacterized serine/threonine-protein kinase SgK494 |
382 |
0.39 |
| chr12_7252075_7252330 | 0.26 |
C1RL |
complement component 1, r subcomponent-like |
2479 |
0.15 |
| chr2_12433970_12434170 | 0.26 |
ENSG00000264089 |
. |
94814 |
0.09 |
| chr4_154448725_154449000 | 0.26 |
KIAA0922 |
KIAA0922 |
28186 |
0.22 |
| chr13_41054978_41055129 | 0.26 |
AL133318.1 |
Uncharacterized protein |
56270 |
0.14 |
| chr7_114566431_114566590 | 0.26 |
MDFIC |
MyoD family inhibitor domain containing |
3379 |
0.39 |
| chr20_57828040_57828198 | 0.26 |
EDN3 |
endothelin 3 |
47363 |
0.15 |
| chr7_797729_797880 | 0.26 |
HEATR2 |
HEAT repeat containing 2 |
10308 |
0.19 |
| chr20_57699560_57699711 | 0.26 |
ZNF831 |
zinc finger protein 831 |
66440 |
0.1 |
| chr3_31219913_31220076 | 0.26 |
ENSG00000265376 |
. |
16715 |
0.26 |
| chr16_79229427_79229578 | 0.26 |
ENSG00000222244 |
. |
68849 |
0.11 |
| chr15_60828599_60828948 | 0.26 |
CTD-2501E16.2 |
|
6601 |
0.21 |
| chr12_9905866_9906017 | 0.26 |
CD69 |
CD69 molecule |
7556 |
0.16 |
| chr2_31360077_31360277 | 0.26 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
710 |
0.75 |
| chr3_49907414_49907654 | 0.25 |
CAMKV |
CaM kinase-like vesicle-associated |
121 |
0.92 |
| chr12_11694203_11694354 | 0.25 |
ENSG00000221128 |
. |
701 |
0.69 |
| chr4_109061717_109062040 | 0.25 |
LEF1 |
lymphoid enhancer-binding factor 1 |
25579 |
0.2 |
| chr10_111846556_111846769 | 0.25 |
ADD3 |
adducin 3 (gamma) |
78940 |
0.09 |
| chr1_172612946_172613385 | 0.25 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
14989 |
0.23 |
| chr8_128751309_128751460 | 0.25 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
2907 |
0.36 |
| chr17_4936150_4936301 | 0.25 |
SLC52A1 |
solute carrier family 52 (riboflavin transporter), member 1 |
2496 |
0.13 |
| chr1_25947708_25947859 | 0.25 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
3442 |
0.25 |
| chr10_134358490_134358742 | 0.25 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
6973 |
0.23 |
| chr12_4257130_4257281 | 0.25 |
CCND2 |
cyclin D2 |
125733 |
0.05 |
| chr11_128591622_128591949 | 0.25 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
25867 |
0.16 |
| chr17_38720137_38720288 | 0.25 |
CCR7 |
chemokine (C-C motif) receptor 7 |
1499 |
0.37 |
| chr17_75964850_75965001 | 0.25 |
TNRC6C |
trinucleotide repeat containing 6C |
35324 |
0.15 |
| chr1_172673559_172674098 | 0.25 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
45670 |
0.18 |
| chr17_32291882_32292036 | 0.25 |
RP11-17M24.2 |
|
11591 |
0.2 |
| chr2_198108993_198109144 | 0.25 |
ANKRD44 |
ankyrin repeat domain 44 |
46306 |
0.12 |
| chr5_6485628_6485779 | 0.25 |
UBE2QL1 |
ubiquitin-conjugating enzyme E2Q family-like 1 |
36967 |
0.21 |
| chr2_42533183_42533346 | 0.25 |
EML4 |
echinoderm microtubule associated protein like 4 |
4871 |
0.28 |
| chr20_18269547_18269794 | 0.25 |
ZNF133 |
zinc finger protein 133 |
492 |
0.7 |
| chr9_92070239_92070498 | 0.25 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
500 |
0.85 |
| chr13_100024268_100024596 | 0.25 |
ENSG00000207719 |
. |
16047 |
0.2 |
| chr6_41990355_41990549 | 0.25 |
ENSG00000206875 |
. |
4200 |
0.19 |
| chr14_71788192_71788343 | 0.25 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
159 |
0.64 |
| chr10_73834893_73835167 | 0.25 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
13056 |
0.23 |
| chr17_214026_214483 | 0.24 |
RP11-1260E13.3 |
|
9726 |
0.13 |
| chr22_40333359_40333568 | 0.24 |
GRAP2 |
GRB2-related adaptor protein 2 |
9358 |
0.17 |
| chr3_100113716_100113867 | 0.24 |
LNP1 |
leukemia NUP98 fusion partner 1 |
6246 |
0.2 |
| chr11_118753286_118753582 | 0.24 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
1041 |
0.35 |
| chr18_67616654_67616805 | 0.24 |
CD226 |
CD226 molecule |
1640 |
0.51 |
| chr18_21445460_21445673 | 0.24 |
LAMA3 |
laminin, alpha 3 |
7238 |
0.23 |
| chr6_41993033_41993209 | 0.24 |
ENSG00000206875 |
. |
6869 |
0.17 |
| chr6_34300867_34301037 | 0.24 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
59499 |
0.1 |
| chr8_38133631_38133782 | 0.24 |
RP11-513D5.5 |
|
300 |
0.85 |
| chr20_31263204_31263355 | 0.24 |
COMMD7 |
COMM domain containing 7 |
67933 |
0.08 |
| chr22_36555180_36555331 | 0.24 |
APOL3 |
apolipoprotein L, 3 |
1576 |
0.43 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.3 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.1 | 0.2 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.2 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.3 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.2 | GO:0002860 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0002837 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.1 | GO:2000758 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0042663 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:1904936 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.2 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.0 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of humoral immune response(GO:0002921) negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.0 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.8 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0072177 | mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0048194 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.0 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0035268 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0030146 | obsolete diuresis(GO:0030146) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.0 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:2000257 | regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.0 | GO:0051971 | positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0031507 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.0 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.0 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.1 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.0 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0032838 | cell projection cytoplasm(GO:0032838) |
| 0.0 | 0.4 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.0 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.4 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.0 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.0 | GO:0036126 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.0 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.2 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0061630 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0016160 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.4 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004938 | alpha-adrenergic receptor activity(GO:0004936) alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.0 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.0 | 0.2 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.0 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |