Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
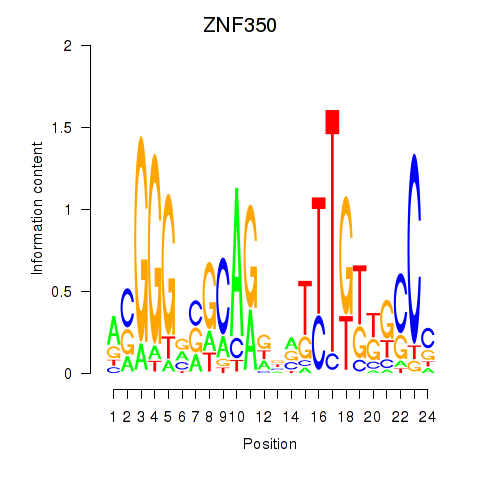
Results for ZNF350
Z-value: 0.75
Transcription factors associated with ZNF350
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF350
|
ENSG00000256683.2 | zinc finger protein 350 |
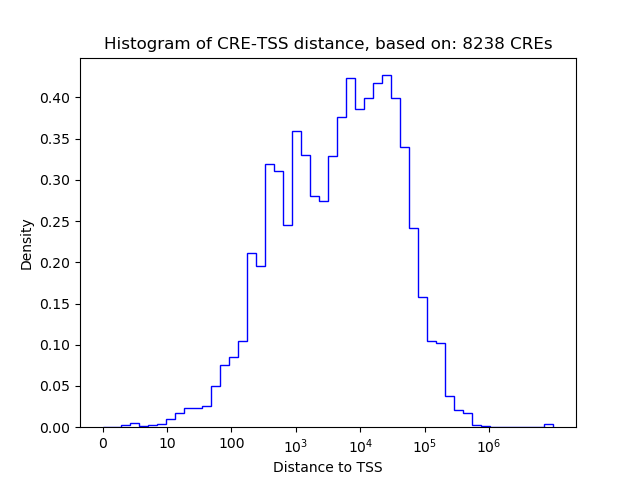
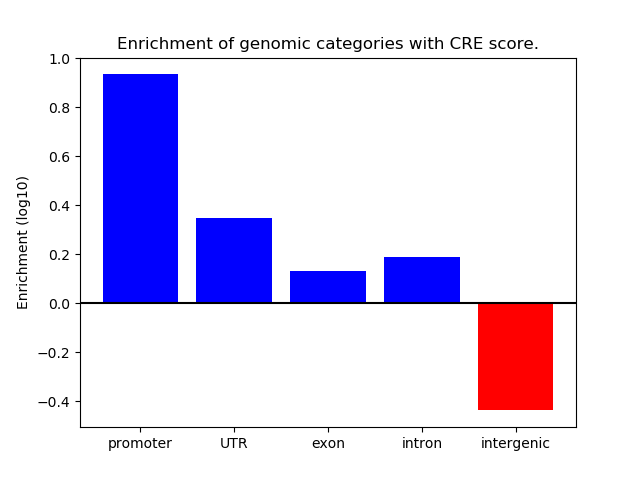
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_52490280_52490507 | ZNF350 | 284 | 0.854308 | 0.69 | 4.1e-02 | Click! |
| chr19_52489445_52489599 | ZNF350 | 401 | 0.770789 | 0.38 | 3.1e-01 | Click! |
| chr19_52489788_52489939 | ZNF350 | 60 | 0.957996 | 0.28 | 4.7e-01 | Click! |
Activity of the ZNF350 motif across conditions
Conditions sorted by the z-value of the ZNF350 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
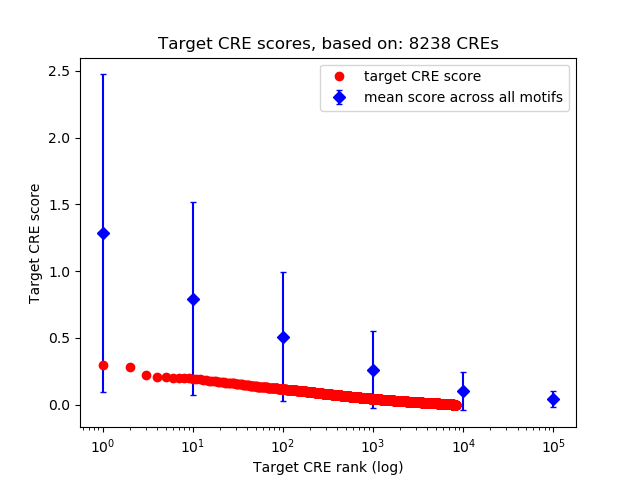
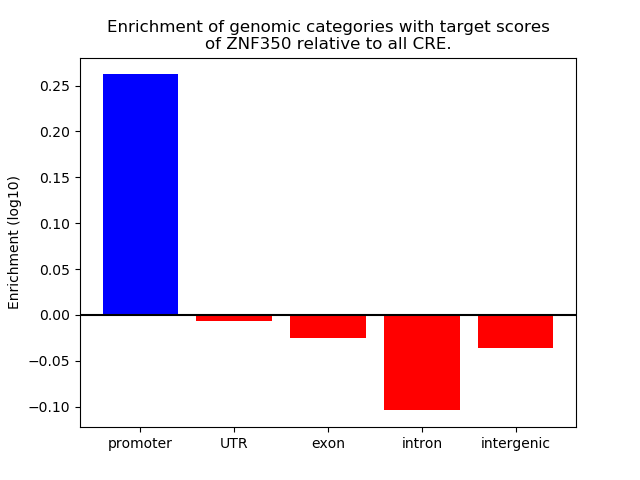
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_79280672_79280844 | 0.30 |
RP11-16K12.1 |
|
9527 |
0.18 |
| chr16_11089953_11090104 | 0.28 |
CLEC16A |
C-type lectin domain family 16, member A |
51590 |
0.09 |
| chr6_41736234_41736385 | 0.22 |
FRS3 |
fibroblast growth factor receptor substrate 3 |
9610 |
0.1 |
| chr9_137349078_137349350 | 0.21 |
RXRA |
retinoid X receptor, alpha |
50786 |
0.15 |
| chr12_25433175_25433326 | 0.21 |
KRAS |
Kirsten rat sarcoma viral oncogene homolog |
29380 |
0.18 |
| chr7_100182794_100183059 | 0.20 |
LRCH4 |
leucine-rich repeats and calponin homology (CH) domain containing 4 |
183 |
0.83 |
| chr1_201719762_201719984 | 0.20 |
NAV1 |
neuron navigator 1 |
10877 |
0.12 |
| chr21_32715120_32715271 | 0.20 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
1399 |
0.58 |
| chr5_14146856_14147066 | 0.20 |
TRIO |
trio Rho guanine nucleotide exchange factor |
3132 |
0.4 |
| chr17_80929713_80929864 | 0.20 |
RP11-1197K16.2 |
|
6613 |
0.21 |
| chr1_113243152_113243493 | 0.19 |
RP11-426L16.3 |
|
4079 |
0.12 |
| chr2_234608672_234608837 | 0.19 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
6416 |
0.1 |
| chr9_124498089_124498240 | 0.19 |
DAB2IP |
DAB2 interacting protein |
6900 |
0.28 |
| chr11_12863782_12864036 | 0.18 |
RP11-47J17.1 |
|
6433 |
0.2 |
| chr20_62686749_62686900 | 0.18 |
RP13-152O15.5 |
|
1332 |
0.22 |
| chr11_69257205_69257356 | 0.18 |
MYEOV |
myeloma overexpressed |
195655 |
0.02 |
| chr20_44978896_44979101 | 0.18 |
SLC35C2 |
solute carrier family 35 (GDP-fucose transporter), member C2 |
8147 |
0.2 |
| chr2_220326118_220326333 | 0.18 |
SPEG |
SPEG complex locus |
118 |
0.93 |
| chr2_20795415_20795848 | 0.17 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
3323 |
0.28 |
| chr20_24929872_24930023 | 0.17 |
CST7 |
cystatin F (leukocystatin) |
81 |
0.97 |
| chr15_70669550_70669810 | 0.17 |
ENSG00000200216 |
. |
184105 |
0.03 |
| chr22_39548986_39549210 | 0.17 |
CBX7 |
chromobox homolog 7 |
443 |
0.8 |
| chr11_63381470_63381652 | 0.16 |
PLA2G16 |
phospholipase A2, group XVI |
280 |
0.86 |
| chr12_124781019_124781262 | 0.16 |
FAM101A |
family with sequence similarity 101, member A |
1071 |
0.61 |
| chr9_72659789_72660086 | 0.16 |
MAMDC2 |
MAM domain containing 2 |
1440 |
0.51 |
| chr1_184005215_184005387 | 0.16 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
1310 |
0.52 |
| chr2_217075636_217075933 | 0.16 |
AC012513.6 |
|
48534 |
0.14 |
| chr7_73120336_73120487 | 0.16 |
ENSG00000265724 |
. |
5236 |
0.11 |
| chr3_156806581_156806732 | 0.16 |
ENSG00000201778 |
. |
64681 |
0.09 |
| chr1_182360343_182360566 | 0.16 |
GLUL |
glutamate-ammonia ligase |
85 |
0.97 |
| chr20_60933838_60933989 | 0.16 |
RP11-157P1.5 |
|
5844 |
0.13 |
| chr19_47407713_47407864 | 0.16 |
ARHGAP35 |
Rho GTPase activating protein 35 |
14145 |
0.17 |
| chr2_160013505_160013656 | 0.16 |
WDSUB1 |
WD repeat, sterile alpha motif and U-box domain containing 1 |
129483 |
0.05 |
| chr9_99482773_99482924 | 0.16 |
ZNF510 |
zinc finger protein 510 |
57480 |
0.13 |
| chr10_131330632_131330830 | 0.15 |
AL355531.2 |
Uncharacterized protein |
21206 |
0.22 |
| chr10_43249103_43249301 | 0.15 |
BMS1 |
BMS1 ribosome biogenesis factor |
29047 |
0.19 |
| chr3_57199098_57199350 | 0.15 |
IL17RD |
interleukin 17 receptor D |
179 |
0.95 |
| chr1_181014307_181014479 | 0.15 |
MR1 |
major histocompatibility complex, class I-related |
11254 |
0.18 |
| chr15_31494061_31494212 | 0.15 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
40660 |
0.18 |
| chr1_113244311_113244462 | 0.15 |
RHOC |
ras homolog family member C |
3157 |
0.13 |
| chr6_166874425_166874576 | 0.15 |
RPS6KA2-IT1 |
RPS6KA2 intronic transcript 1 (non-protein coding) |
4371 |
0.23 |
| chr13_113583857_113584008 | 0.15 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
27423 |
0.15 |
| chr11_128774089_128774256 | 0.14 |
C11orf45 |
chromosome 11 open reading frame 45 |
1420 |
0.38 |
| chr7_17234854_17235017 | 0.14 |
AC003075.4 |
|
85992 |
0.1 |
| chr4_1037278_1037479 | 0.14 |
FGFRL1 |
fibroblast growth factor receptor-like 1 |
31139 |
0.11 |
| chr5_673661_673812 | 0.14 |
CTD-2589H19.6 |
|
2205 |
0.24 |
| chr14_74984739_74984890 | 0.14 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
8304 |
0.16 |
| chr17_55131688_55131839 | 0.14 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
30690 |
0.13 |
| chr14_93043446_93043626 | 0.14 |
RIN3 |
Ras and Rab interactor 3 |
63388 |
0.13 |
| chr18_20772119_20772404 | 0.14 |
CABLES1 |
Cdk5 and Abl enzyme substrate 1 |
36472 |
0.16 |
| chr9_131936536_131936750 | 0.14 |
RP11-247A12.8 |
|
1443 |
0.27 |
| chr11_66650721_66650872 | 0.14 |
PC |
pyruvate carboxylase |
24576 |
0.11 |
| chr15_70877190_70877341 | 0.14 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
117355 |
0.06 |
| chr16_11348403_11348570 | 0.14 |
SOCS1 |
suppressor of cytokine signaling 1 |
1550 |
0.21 |
| chr2_87901921_87902194 | 0.14 |
ENSG00000265507 |
. |
27217 |
0.24 |
| chr8_37552274_37552425 | 0.14 |
ZNF703 |
zinc finger protein 703 |
920 |
0.47 |
| chr1_3620112_3620445 | 0.13 |
TP73 |
tumor protein p73 |
5656 |
0.13 |
| chr11_66823005_66823323 | 0.13 |
RHOD |
ras homolog family member D |
1125 |
0.44 |
| chr3_169939455_169939857 | 0.13 |
PRKCI |
protein kinase C, iota |
497 |
0.79 |
| chr3_13323123_13323361 | 0.13 |
NUP210 |
nucleoporin 210kDa |
138567 |
0.05 |
| chr19_36359879_36360032 | 0.13 |
APLP1 |
amyloid beta (A4) precursor-like protein 1 |
420 |
0.67 |
| chr3_14494260_14494411 | 0.13 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
19415 |
0.22 |
| chr1_22236518_22236840 | 0.13 |
HSPG2 |
heparan sulfate proteoglycan 2 |
13876 |
0.16 |
| chr10_115827772_115827923 | 0.13 |
ADRB1 |
adrenoceptor beta 1 |
24041 |
0.18 |
| chr3_62573282_62573433 | 0.13 |
CADPS |
Ca++-dependent secretion activator |
2436 |
0.44 |
| chr6_71666612_71666763 | 0.13 |
B3GAT2 |
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
54 |
0.99 |
| chr1_54764314_54764483 | 0.13 |
RP5-997D24.3 |
|
13320 |
0.18 |
| chr19_39423223_39423374 | 0.13 |
MRPS12 |
mitochondrial ribosomal protein S12 |
1659 |
0.17 |
| chr6_135504457_135504736 | 0.13 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
1944 |
0.33 |
| chr1_154980745_154981008 | 0.13 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
5581 |
0.08 |
| chr17_7119350_7119780 | 0.13 |
ACADVL |
acyl-CoA dehydrogenase, very long chain |
879 |
0.26 |
| chr3_56915864_56916077 | 0.13 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
34529 |
0.19 |
| chr17_4693016_4693203 | 0.13 |
GLTPD2 |
glycolipid transfer protein domain containing 2 |
855 |
0.36 |
| chr2_73151273_73151429 | 0.13 |
EMX1 |
empty spiracles homeobox 1 |
234 |
0.94 |
| chr8_59571402_59571553 | 0.13 |
NSMAF |
neutral sphingomyelinase (N-SMase) activation associated factor |
616 |
0.76 |
| chr8_31497191_31497379 | 0.13 |
NRG1 |
neuregulin 1 |
14 |
0.99 |
| chr4_75858432_75858583 | 0.13 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
181 |
0.96 |
| chr2_112105742_112105987 | 0.13 |
ENSG00000266139 |
. |
27196 |
0.26 |
| chr5_140887603_140887841 | 0.13 |
DIAPH1 |
diaphanous-related formin 1 |
17992 |
0.08 |
| chr1_151483317_151483639 | 0.13 |
CGN |
cingulin |
66 |
0.95 |
| chr15_91210216_91210381 | 0.13 |
RP11-387D10.3 |
|
6328 |
0.17 |
| chr19_36398732_36398883 | 0.12 |
TYROBP |
TYRO protein tyrosine kinase binding protein |
342 |
0.75 |
| chr15_59739965_59740116 | 0.12 |
FAM81A |
family with sequence similarity 81, member A |
2735 |
0.25 |
| chr17_33570615_33570828 | 0.12 |
SLFN5 |
schlafen family member 5 |
613 |
0.47 |
| chr2_95941109_95941447 | 0.12 |
PROM2 |
prominin 2 |
1033 |
0.58 |
| chr16_73191000_73191151 | 0.12 |
C16orf47 |
chromosome 16 open reading frame 47 |
12729 |
0.28 |
| chr4_186065139_186065290 | 0.12 |
SLC25A4 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
819 |
0.63 |
| chr9_139492984_139493135 | 0.12 |
ENSG00000252440 |
. |
3908 |
0.13 |
| chr11_69251734_69251885 | 0.12 |
MYEOV |
myeloma overexpressed |
190184 |
0.03 |
| chr11_34267398_34267549 | 0.12 |
ENSG00000201867 |
. |
56752 |
0.14 |
| chr17_39940723_39940965 | 0.12 |
JUP |
junction plakoglobin |
563 |
0.6 |
| chr4_140871909_140872439 | 0.12 |
MAML3 |
mastermind-like 3 (Drosophila) |
60053 |
0.14 |
| chr22_50707294_50707445 | 0.12 |
MAPK11 |
mitogen-activated protein kinase 11 |
1424 |
0.21 |
| chr14_100003657_100003808 | 0.12 |
CCDC85C |
coiled-coil domain containing 85C |
1307 |
0.41 |
| chr1_27917666_27917818 | 0.12 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
12360 |
0.15 |
| chr5_139046723_139047090 | 0.12 |
CXXC5 |
CXXC finger protein 5 |
7766 |
0.22 |
| chr17_38698835_38699076 | 0.12 |
CCR7 |
chemokine (C-C motif) receptor 7 |
18310 |
0.15 |
| chr5_175970644_175970798 | 0.12 |
CDHR2 |
cadherin-related family member 2 |
1209 |
0.33 |
| chr1_153517829_153518044 | 0.12 |
S100A4 |
S100 calcium binding protein A4 |
342 |
0.73 |
| chr20_10551784_10551935 | 0.12 |
JAG1 |
jagged 1 |
91295 |
0.08 |
| chr6_90836574_90836725 | 0.12 |
ENSG00000222078 |
. |
125424 |
0.05 |
| chr17_79065717_79065990 | 0.12 |
BAIAP2 |
BAI1-associated protein 2 |
5526 |
0.12 |
| chr16_55535078_55535229 | 0.12 |
LPCAT2 |
lysophosphatidylcholine acyltransferase 2 |
7757 |
0.22 |
| chr9_133641708_133641929 | 0.12 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
52451 |
0.13 |
| chr14_63861661_63861812 | 0.12 |
ENSG00000264995 |
. |
67988 |
0.1 |
| chr15_74248873_74249108 | 0.12 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
28401 |
0.11 |
| chr21_36250882_36251033 | 0.11 |
RUNX1 |
runt-related transcription factor 1 |
8523 |
0.31 |
| chr8_21313579_21313730 | 0.11 |
ENSG00000266713 |
. |
37885 |
0.21 |
| chr10_49816492_49816643 | 0.11 |
ARHGAP22 |
Rho GTPase activating protein 22 |
3429 |
0.27 |
| chr1_20959712_20959863 | 0.11 |
PINK1 |
PTEN induced putative kinase 1 |
161 |
0.95 |
| chr2_133195967_133196118 | 0.11 |
GPR39 |
G protein-coupled receptor 39 |
21895 |
0.19 |
| chr17_56409271_56409708 | 0.11 |
MIR142 |
microRNA 142 |
380 |
0.74 |
| chr3_193491368_193491519 | 0.11 |
ENSG00000243991 |
. |
32933 |
0.21 |
| chr9_90115986_90116137 | 0.11 |
DAPK1 |
death-associated protein kinase 1 |
2141 |
0.41 |
| chr1_156553418_156553569 | 0.11 |
TTC24 |
tetratricopeptide repeat domain 24 |
2336 |
0.14 |
| chr8_97506971_97507442 | 0.11 |
SDC2 |
syndecan 2 |
970 |
0.67 |
| chr4_56501418_56501774 | 0.11 |
NMU |
neuromedin U |
830 |
0.65 |
| chr1_95201035_95201215 | 0.11 |
ENSG00000263526 |
. |
10331 |
0.26 |
| chr16_31342390_31342554 | 0.11 |
ITGAM |
integrin, alpha M (complement component 3 receptor 3 subunit) |
771 |
0.53 |
| chr19_1131499_1131650 | 0.11 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
646 |
0.58 |
| chr9_132090472_132090833 | 0.11 |
ENSG00000242281 |
. |
42088 |
0.14 |
| chr8_125388828_125388979 | 0.11 |
TMEM65 |
transmembrane protein 65 |
3970 |
0.28 |
| chr12_54426850_54427001 | 0.11 |
HOXC5 |
homeobox C5 |
288 |
0.68 |
| chr5_138474799_138474993 | 0.11 |
SIL1 |
SIL1 nucleotide exchange factor |
7449 |
0.22 |
| chr19_48011511_48011767 | 0.11 |
NAPA |
N-ethylmaleimide-sensitive factor attachment protein, alpha |
4384 |
0.15 |
| chr12_56242031_56242182 | 0.11 |
MMP19 |
matrix metallopeptidase 19 |
5372 |
0.1 |
| chr12_53342557_53342709 | 0.11 |
KRT18 |
keratin 18 |
22 |
0.94 |
| chr9_131903199_131903823 | 0.11 |
PPP2R4 |
protein phosphatase 2A activator, regulatory subunit 4 |
414 |
0.76 |
| chr12_122239357_122239755 | 0.11 |
RHOF |
ras homolog family member F (in filopodia) |
973 |
0.47 |
| chr1_152022964_152023546 | 0.11 |
S100A11 |
S100 calcium binding protein A11 |
13744 |
0.14 |
| chr20_25018363_25018514 | 0.11 |
ACSS1 |
acyl-CoA synthetase short-chain family member 1 |
5099 |
0.22 |
| chr8_22779071_22779222 | 0.11 |
PEBP4 |
phosphatidylethanolamine-binding protein 4 |
6275 |
0.16 |
| chr6_33806569_33806720 | 0.11 |
MLN |
motilin |
34856 |
0.15 |
| chr9_127056098_127056249 | 0.11 |
NEK6 |
NIMA-related kinase 6 |
1474 |
0.4 |
| chr5_109400140_109400292 | 0.11 |
AC011366.3 |
Uncharacterized protein |
181333 |
0.03 |
| chr11_67908129_67908418 | 0.11 |
CTD-2655K5.1 |
|
11008 |
0.15 |
| chr20_58586927_58587156 | 0.11 |
CDH26 |
cadherin 26 |
15592 |
0.2 |
| chr16_30007199_30007350 | 0.11 |
INO80E |
INO80 complex subunit E |
238 |
0.53 |
| chr10_63546087_63546274 | 0.11 |
RP11-63A2.2 |
|
101034 |
0.07 |
| chr9_127058687_127058838 | 0.11 |
NEK6 |
NIMA-related kinase 6 |
3502 |
0.23 |
| chr11_133904571_133904867 | 0.11 |
JAM3 |
junctional adhesion molecule 3 |
34101 |
0.17 |
| chr22_35874169_35874336 | 0.11 |
RASD2 |
RASD family, member 2 |
62663 |
0.11 |
| chr21_38071422_38071589 | 0.11 |
SIM2 |
single-minded family bHLH transcription factor 2 |
71 |
0.97 |
| chr15_41245906_41246096 | 0.11 |
CHAC1 |
ChaC, cation transport regulator homolog 1 (E. coli) |
403 |
0.79 |
| chr19_6239081_6239341 | 0.11 |
CTC-503J8.4 |
|
19957 |
0.13 |
| chr5_176922042_176922394 | 0.11 |
RP11-1334A24.6 |
|
222 |
0.85 |
| chr17_74488583_74488734 | 0.11 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
578 |
0.6 |
| chr7_23312941_23313332 | 0.10 |
MALSU1 |
mitochondrial assembly of ribosomal large subunit 1 |
25683 |
0.17 |
| chr1_16861633_16861784 | 0.10 |
ENSG00000233421 |
. |
989 |
0.41 |
| chr14_103590026_103590426 | 0.10 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
428 |
0.82 |
| chr2_217529680_217529831 | 0.10 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
4299 |
0.2 |
| chr22_35937290_35937443 | 0.10 |
RASD2 |
RASD family, member 2 |
451 |
0.86 |
| chr2_88901946_88902210 | 0.10 |
EIF2AK3 |
eukaryotic translation initiation factor 2-alpha kinase 3 |
6703 |
0.18 |
| chr11_66428802_66428953 | 0.10 |
RP11-658F2.8 |
|
4630 |
0.11 |
| chr9_14348782_14348933 | 0.10 |
RP11-120J1.1 |
|
1538 |
0.5 |
| chr6_125420749_125420900 | 0.10 |
TPD52L1 |
tumor protein D52-like 1 |
19371 |
0.24 |
| chr2_129493191_129493342 | 0.10 |
ENSG00000238379 |
. |
290494 |
0.01 |
| chr17_46020955_46021139 | 0.10 |
PNPO |
pyridoxamine 5'-phosphate oxidase |
2056 |
0.15 |
| chr1_3511510_3511661 | 0.10 |
MEGF6 |
multiple EGF-like-domains 6 |
16474 |
0.13 |
| chr14_105147633_105147901 | 0.10 |
ENSG00000265291 |
. |
3681 |
0.16 |
| chr12_97379822_97380067 | 0.10 |
ENSG00000202368 |
. |
6537 |
0.31 |
| chr5_139154506_139154831 | 0.10 |
PSD2 |
pleckstrin and Sec7 domain containing 2 |
20738 |
0.19 |
| chr16_85989408_85989698 | 0.10 |
IRF8 |
interferon regulatory factor 8 |
41634 |
0.18 |
| chr9_124398001_124398152 | 0.10 |
DAB2IP |
DAB2 interacting protein |
15797 |
0.23 |
| chr3_58044055_58044466 | 0.10 |
FLNB |
filamin B, beta |
19796 |
0.24 |
| chr22_25373670_25373970 | 0.10 |
KIAA1671 |
KIAA1671 |
25123 |
0.18 |
| chr19_38517988_38518282 | 0.10 |
ENSG00000221258 |
. |
26470 |
0.15 |
| chr10_112261658_112261918 | 0.10 |
DUSP5 |
dual specificity phosphatase 5 |
4192 |
0.22 |
| chr4_157996843_157997211 | 0.10 |
GLRB |
glycine receptor, beta |
182 |
0.96 |
| chr2_242714597_242714748 | 0.10 |
GAL3ST2 |
galactose-3-O-sulfotransferase 2 |
1568 |
0.23 |
| chr16_57418029_57418329 | 0.10 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
7295 |
0.14 |
| chr11_69453616_69453767 | 0.10 |
CCND1 |
cyclin D1 |
2164 |
0.35 |
| chr2_227663253_227663404 | 0.10 |
IRS1 |
insulin receptor substrate 1 |
1147 |
0.51 |
| chr14_103991038_103991189 | 0.10 |
CKB |
creatine kinase, brain |
1665 |
0.19 |
| chr20_35990779_35990930 | 0.10 |
SRC |
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
16297 |
0.23 |
| chr12_53626909_53627429 | 0.10 |
RARG |
retinoic acid receptor, gamma |
405 |
0.74 |
| chr1_25070262_25070509 | 0.10 |
CLIC4 |
chloride intracellular channel 4 |
1463 |
0.44 |
| chr10_29702503_29702654 | 0.10 |
PTCHD3P1 |
patched domain containing 3 pseudogene 1 |
3406 |
0.33 |
| chr1_25321403_25321690 | 0.10 |
ENSG00000264371 |
. |
28448 |
0.16 |
| chr9_140329267_140329418 | 0.10 |
ENTPD8 |
ectonucleoside triphosphate diphosphohydrolase 8 |
3356 |
0.14 |
| chr20_4666254_4666846 | 0.10 |
PRNP |
prion protein |
332 |
0.91 |
| chr3_42567586_42567737 | 0.10 |
VIPR1-AS1 |
VIPR1 antisense RNA 1 |
5893 |
0.16 |
| chr1_42136653_42136804 | 0.10 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
29944 |
0.22 |
| chr13_100073797_100073948 | 0.10 |
ENSG00000266207 |
. |
31851 |
0.16 |
| chr3_129368977_129369267 | 0.10 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
6447 |
0.21 |
| chr1_236444541_236444706 | 0.10 |
ERO1LB |
ERO1-like beta (S. cerevisiae) |
278 |
0.91 |
| chr17_80846919_80847070 | 0.10 |
TBCD |
tubulin folding cofactor D |
3034 |
0.26 |
| chr13_100070069_100070311 | 0.10 |
ENSG00000266207 |
. |
35533 |
0.15 |
| chr10_49496390_49496541 | 0.10 |
FRMPD2 |
FERM and PDZ domain containing 2 |
13524 |
0.22 |
| chr2_231741597_231741748 | 0.10 |
AC012507.3 |
|
9543 |
0.16 |
| chr17_42163716_42163867 | 0.10 |
HDAC5 |
histone deacetylase 5 |
6764 |
0.1 |
| chr1_153463963_153464114 | 0.10 |
ENSG00000263841 |
. |
9202 |
0.1 |
| chr14_102974651_102974824 | 0.10 |
ANKRD9 |
ankyrin repeat domain 9 |
417 |
0.82 |
| chr9_116383050_116383201 | 0.10 |
RGS3 |
regulator of G-protein signaling 3 |
27359 |
0.19 |
| chr3_135915447_135915650 | 0.10 |
MSL2 |
male-specific lethal 2 homolog (Drosophila) |
117 |
0.98 |
| chr8_95961948_95962152 | 0.10 |
RP11-347C18.3 |
|
215 |
0.69 |
| chr15_81072143_81072333 | 0.10 |
KIAA1199 |
KIAA1199 |
526 |
0.82 |
| chr11_2011512_2011663 | 0.10 |
MRPL23-AS1 |
MRPL23 antisense RNA 1 |
437 |
0.77 |
| chr12_123546608_123546759 | 0.10 |
PITPNM2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
13959 |
0.16 |
| chr1_204126949_204127100 | 0.09 |
ETNK2 |
ethanolamine kinase 2 |
5717 |
0.14 |
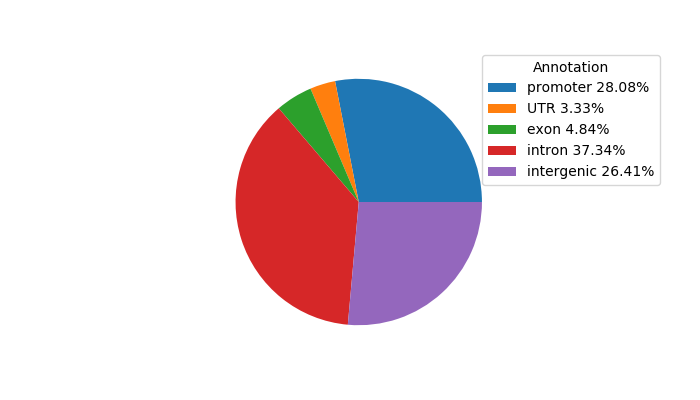
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0003170 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.0 | GO:0061037 | negative regulation of cartilage development(GO:0061037) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.2 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.0 | 0.0 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0033505 | floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0060856 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.1 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.0 | 0.0 | GO:0061213 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0003416 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0071803 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0051532 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.0 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.0 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |