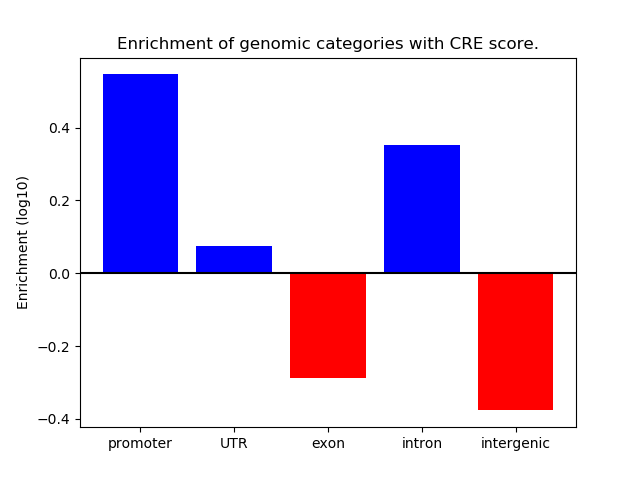
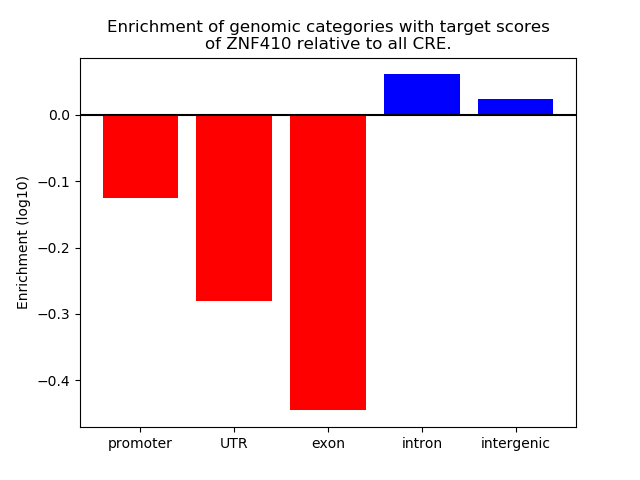
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
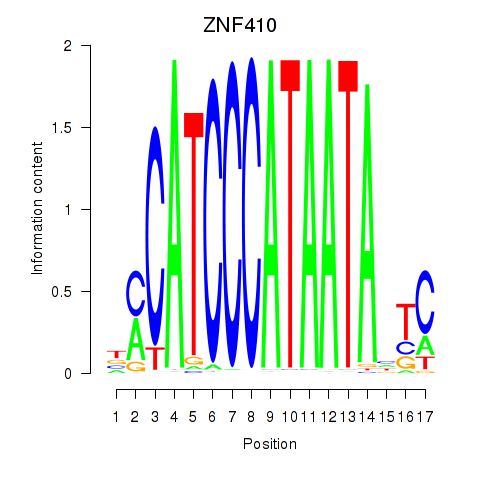
Results for ZNF410
Z-value: 0.50
Transcription factors associated with ZNF410
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF410
|
ENSG00000119725.13 | zinc finger protein 410 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_74366133_74366284 | ZNF410 | 4195 | 0.118388 | 0.68 | 4.4e-02 | Click! |
| chr14_74366502_74366653 | ZNF410 | 3826 | 0.122332 | 0.63 | 6.6e-02 | Click! |
| chr14_74352029_74352386 | ZNF410 | 1113 | 0.340434 | -0.57 | 1.1e-01 | Click! |
| chr14_74355053_74355205 | ZNF410 | 1368 | 0.274815 | 0.43 | 2.5e-01 | Click! |
| chr14_74356161_74356312 | ZNF410 | 2317 | 0.170419 | 0.40 | 2.8e-01 | Click! |
Activity of the ZNF410 motif across conditions
Conditions sorted by the z-value of the ZNF410 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
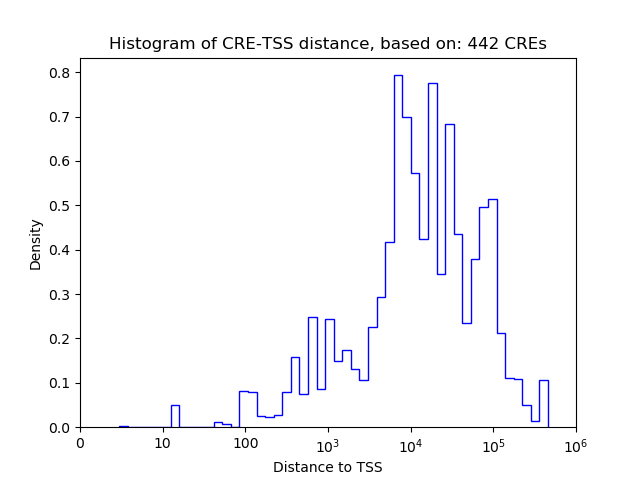
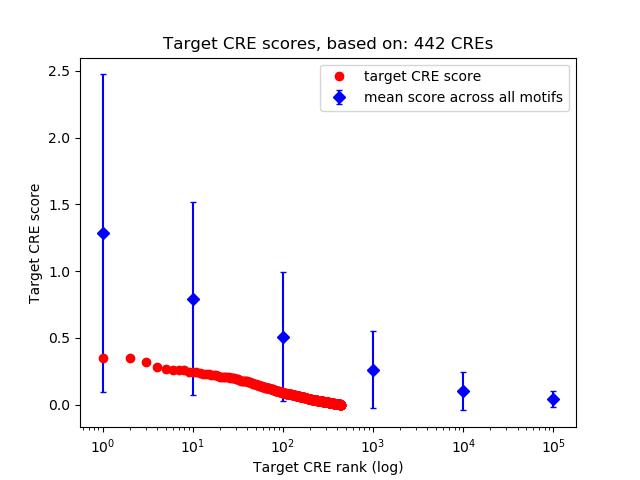
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_6716949_6717188 | 0.35 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
426 |
0.66 |
| chr12_6717224_6717396 | 0.35 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
668 |
0.47 |
| chr3_99801308_99801543 | 0.32 |
FILIP1L |
filamin A interacting protein 1-like |
31932 |
0.17 |
| chr2_56115946_56116097 | 0.28 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
34335 |
0.17 |
| chr11_15179125_15179276 | 0.27 |
INSC |
inscuteable homolog (Drosophila) |
8546 |
0.3 |
| chr3_30538477_30538630 | 0.26 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
109441 |
0.07 |
| chr2_33329316_33329467 | 0.26 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
30082 |
0.23 |
| chr6_140372368_140372519 | 0.26 |
ENSG00000252107 |
. |
107388 |
0.08 |
| chr5_88733052_88733203 | 0.24 |
MEF2C-AS1 |
MEF2C antisense RNA 1 |
18503 |
0.3 |
| chr15_58734161_58734312 | 0.24 |
LIPC |
lipase, hepatic |
10046 |
0.2 |
| chr3_40151249_40151400 | 0.24 |
MYRIP |
myosin VIIA and Rab interacting protein |
9822 |
0.27 |
| chr11_74561231_74561382 | 0.24 |
ENSG00000242999 |
. |
4351 |
0.2 |
| chr12_29196725_29196876 | 0.23 |
ENSG00000222481 |
. |
77015 |
0.11 |
| chr7_77200209_77200424 | 0.23 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
92 |
0.98 |
| chr10_111659026_111659177 | 0.23 |
XPNPEP1 |
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
8476 |
0.22 |
| chr8_30309779_30309930 | 0.23 |
RBPMS |
RNA binding protein with multiple splicing |
9721 |
0.26 |
| chr16_53246810_53247380 | 0.23 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
4732 |
0.24 |
| chr12_81172448_81172599 | 0.22 |
ENSG00000207763 |
. |
53885 |
0.12 |
| chr12_46776255_46776796 | 0.22 |
SLC38A2 |
solute carrier family 38, member 2 |
9875 |
0.24 |
| chr18_20945112_20945263 | 0.21 |
TMEM241 |
transmembrane protein 241 |
72630 |
0.09 |
| chr1_68306464_68306615 | 0.21 |
GNG12 |
guanine nucleotide binding protein (G protein), gamma 12 |
7389 |
0.22 |
| chr5_9194903_9195054 | 0.21 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
70679 |
0.13 |
| chr1_55012899_55013050 | 0.21 |
ACOT11 |
acyl-CoA thioesterase 11 |
927 |
0.55 |
| chr9_87838634_87838879 | 0.21 |
AGTPBP1 |
ATP/GTP binding protein 1 |
457445 |
0.01 |
| chr3_190245134_190245536 | 0.21 |
IL1RAP |
interleukin 1 receptor accessory protein |
13444 |
0.26 |
| chr17_54262776_54262927 | 0.20 |
ANKFN1 |
ankyrin-repeat and fibronectin type III domain containing 1 |
11315 |
0.31 |
| chr1_33629505_33629812 | 0.20 |
TRIM62 |
tripartite motif containing 62 |
12493 |
0.21 |
| chr1_179148308_179148459 | 0.20 |
ENSG00000212338 |
. |
17225 |
0.17 |
| chr8_26206285_26206436 | 0.20 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
34054 |
0.17 |
| chrX_16661091_16661259 | 0.20 |
S100G |
S100 calcium binding protein G |
7106 |
0.18 |
| chr7_34022267_34022418 | 0.20 |
BMPER |
BMP binding endothelial regulator |
77197 |
0.12 |
| chr3_99997916_99998067 | 0.19 |
TBC1D23 |
TBC1 domain family, member 23 |
293 |
0.91 |
| chr11_5385191_5385342 | 0.19 |
OR51B6 |
olfactory receptor, family 51, subfamily B, member 6 |
12528 |
0.1 |
| chr1_204395036_204395187 | 0.18 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
14192 |
0.17 |
| chr1_95219154_95219305 | 0.18 |
ENSG00000263526 |
. |
7773 |
0.26 |
| chr15_40234024_40234175 | 0.18 |
EIF2AK4 |
eukaryotic translation initiation factor 2 alpha kinase 4 |
7722 |
0.15 |
| chr3_57018939_57019090 | 0.18 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
7611 |
0.23 |
| chr1_28466094_28466257 | 0.18 |
ENSG00000253005 |
. |
3177 |
0.17 |
| chr3_101555850_101556001 | 0.18 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
9088 |
0.19 |
| chr13_21654358_21654593 | 0.17 |
LATS2 |
large tumor suppressor kinase 2 |
18789 |
0.14 |
| chr20_31332968_31333119 | 0.17 |
COMMD7 |
COMM domain containing 7 |
1240 |
0.47 |
| chr10_97039069_97039220 | 0.17 |
PDLIM1 |
PDZ and LIM domain 1 |
11637 |
0.23 |
| chr14_29751933_29752084 | 0.17 |
ENSG00000257522 |
. |
95854 |
0.09 |
| chr2_3495267_3495450 | 0.16 |
TRAPPC12-AS1 |
TRAPPC12 antisense RNA 1 |
9178 |
0.14 |
| chr6_157610859_157611010 | 0.16 |
ENSG00000252609 |
. |
101497 |
0.07 |
| chr7_13920146_13920297 | 0.16 |
ETV1 |
ets variant 1 |
105845 |
0.08 |
| chr10_98460620_98460814 | 0.16 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
19554 |
0.19 |
| chr5_14154581_14154957 | 0.16 |
TRIO |
trio Rho guanine nucleotide exchange factor |
10940 |
0.32 |
| chr6_47010736_47010893 | 0.16 |
GPR110 |
G protein-coupled receptor 110 |
715 |
0.78 |
| chr3_115155334_115155485 | 0.16 |
GAP43 |
growth associated protein 43 |
186762 |
0.03 |
| chr14_35097827_35098332 | 0.15 |
SNX6 |
sorting nexin 6 |
799 |
0.64 |
| chr2_221144537_221144688 | 0.15 |
ENSG00000221199 |
. |
88013 |
0.1 |
| chr6_132720652_132720803 | 0.15 |
MOXD1 |
monooxygenase, DBH-like 1 |
1887 |
0.45 |
| chr4_77577306_77577457 | 0.15 |
AC107072.2 |
|
18605 |
0.2 |
| chr5_57591370_57591521 | 0.14 |
PLK2 |
polo-like kinase 2 |
164642 |
0.04 |
| chr2_201612308_201612459 | 0.14 |
ENSG00000201737 |
. |
27924 |
0.13 |
| chr5_36725188_36725339 | 0.14 |
CTD-2353F22.1 |
|
12 |
0.99 |
| chr10_15834401_15834552 | 0.14 |
FAM188A |
family with sequence similarity 188, member A |
25531 |
0.23 |
| chr10_78046565_78046716 | 0.14 |
ENSG00000201954 |
. |
25979 |
0.18 |
| chr16_70031993_70032144 | 0.14 |
RP11-419C5.2 |
|
1977 |
0.28 |
| chr9_117077655_117077806 | 0.14 |
ORM1 |
orosomucoid 1 |
7606 |
0.19 |
| chr2_152213518_152213794 | 0.13 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
450 |
0.8 |
| chr1_211770661_211771027 | 0.13 |
RP11-359E8.5 |
|
13777 |
0.17 |
| chr2_75122030_75122181 | 0.13 |
HK2 |
hexokinase 2 |
59808 |
0.12 |
| chr17_36862672_36863533 | 0.13 |
CTB-58E17.3 |
|
588 |
0.44 |
| chr14_55088387_55088538 | 0.13 |
SAMD4A |
sterile alpha motif domain containing 4A |
53825 |
0.15 |
| chr8_19521264_19521461 | 0.13 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
18732 |
0.28 |
| chr1_229118656_229118807 | 0.12 |
RP5-1061H20.5 |
|
244578 |
0.01 |
| chr6_73298808_73298959 | 0.12 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
32637 |
0.2 |
| chr12_13074467_13074618 | 0.12 |
ENSG00000207817 |
. |
5779 |
0.13 |
| chr4_88727397_88727610 | 0.12 |
IBSP |
integrin-binding sialoprotein |
6770 |
0.18 |
| chr12_6722253_6722786 | 0.12 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
5877 |
0.09 |
| chr20_45928695_45928846 | 0.12 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
1126 |
0.41 |
| chr4_7907002_7907153 | 0.12 |
AFAP1 |
actin filament associated protein 1 |
33037 |
0.16 |
| chr9_80211377_80211528 | 0.12 |
ENSG00000201711 |
. |
38773 |
0.19 |
| chr20_25506429_25506580 | 0.12 |
NINL |
ninein-like |
46841 |
0.14 |
| chr3_156266075_156266226 | 0.12 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
5764 |
0.24 |
| chr13_25485161_25485312 | 0.11 |
CENPJ |
centromere protein J |
7104 |
0.24 |
| chr16_14839837_14839988 | 0.11 |
RP11-719K4.6 |
|
122 |
0.94 |
| chr16_14800717_14800868 | 0.11 |
RP11-82O18.2 |
|
120 |
0.93 |
| chr3_100235412_100235563 | 0.11 |
TMEM45A |
transmembrane protein 45A |
23983 |
0.21 |
| chr6_88457610_88457792 | 0.11 |
ENSG00000238628 |
. |
27068 |
0.17 |
| chr5_142254524_142254675 | 0.11 |
ARHGAP26-AS1 |
ARHGAP26 antisense RNA 1 |
6112 |
0.27 |
| chr5_32841630_32841781 | 0.10 |
AC026703.1 |
|
52760 |
0.15 |
| chr2_15370019_15370258 | 0.10 |
NBAS |
neuroblastoma amplified sequence |
8672 |
0.33 |
| chr10_24753767_24753918 | 0.10 |
KIAA1217 |
KIAA1217 |
1618 |
0.45 |
| chr5_163628955_163629106 | 0.10 |
CTC-340A15.2 |
|
94672 |
0.1 |
| chrX_24073482_24074722 | 0.10 |
EIF2S3 |
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
1014 |
0.57 |
| chr8_125815205_125815550 | 0.10 |
ENSG00000263735 |
. |
18923 |
0.24 |
| chr9_129811608_129811922 | 0.10 |
ANGPTL2 |
angiopoietin-like 2 |
73148 |
0.1 |
| chr8_70091869_70092020 | 0.10 |
ENSG00000238808 |
. |
69135 |
0.14 |
| chr16_81779154_81779305 | 0.10 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
6527 |
0.3 |
| chr8_103938601_103938752 | 0.10 |
ENSG00000201216 |
. |
24116 |
0.13 |
| chrX_123403964_123404115 | 0.10 |
ENSG00000252693 |
. |
72446 |
0.11 |
| chr20_36629780_36630040 | 0.09 |
TTI1 |
TELO2 interacting protein 1 |
12395 |
0.17 |
| chr3_67559756_67559907 | 0.09 |
SUCLG2 |
succinate-CoA ligase, GDP-forming, beta subunit |
18815 |
0.3 |
| chr12_6717643_6717941 | 0.09 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
1150 |
0.27 |
| chr17_35854791_35854942 | 0.09 |
DUSP14 |
dual specificity phosphatase 14 |
3296 |
0.25 |
| chr7_133373225_133373376 | 0.09 |
EXOC4 |
exocyst complex component 4 |
112084 |
0.07 |
| chr10_103608566_103608725 | 0.09 |
KCNIP2 |
Kv channel interacting protein 2 |
4968 |
0.18 |
| chr11_109983287_109983623 | 0.09 |
ZC3H12C |
zinc finger CCCH-type containing 12C |
18274 |
0.29 |
| chr1_42312204_42312355 | 0.09 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
71890 |
0.12 |
| chr6_21610679_21610963 | 0.09 |
SOX4 |
SRY (sex determining region Y)-box 4 |
16750 |
0.31 |
| chr2_1629536_1629687 | 0.09 |
AC144450.1 |
|
5726 |
0.29 |
| chr1_159065750_159065901 | 0.09 |
AIM2 |
absent in melanoma 2 |
19134 |
0.17 |
| chr5_39460419_39460570 | 0.09 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
1908 |
0.49 |
| chr15_101726120_101726271 | 0.08 |
CHSY1 |
chondroitin sulfate synthase 1 |
65942 |
0.1 |
| chr7_94327256_94327407 | 0.08 |
ENSG00000239030 |
. |
14390 |
0.2 |
| chr17_1356696_1356953 | 0.08 |
CRK |
v-crk avian sarcoma virus CT10 oncogene homolog |
2619 |
0.21 |
| chr9_132977954_132978121 | 0.08 |
NCS1 |
neuronal calcium sensor 1 |
15165 |
0.22 |
| chr17_73872544_73873300 | 0.08 |
TRIM47 |
tripartite motif containing 47 |
423 |
0.43 |
| chr1_36751423_36751675 | 0.08 |
SH3D21 |
SH3 domain containing 21 |
20439 |
0.13 |
| chr1_212421007_212421158 | 0.08 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
37797 |
0.14 |
| chr1_150188550_150188701 | 0.08 |
ENSG00000266187 |
. |
4695 |
0.12 |
| chr1_84764836_84765174 | 0.08 |
SAMD13 |
sterile alpha motif domain containing 13 |
236 |
0.94 |
| chr6_147534808_147534959 | 0.08 |
STXBP5 |
syntaxin binding protein 5 (tomosyn) |
7773 |
0.25 |
| chr3_71574289_71574598 | 0.08 |
ENSG00000221264 |
. |
16797 |
0.2 |
| chr10_30573247_30573398 | 0.08 |
ENSG00000200887 |
. |
15280 |
0.19 |
| chr6_135857036_135857769 | 0.08 |
AHI1 |
Abelson helper integration site 1 |
38488 |
0.19 |
| chr5_141695794_141696287 | 0.08 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
7707 |
0.23 |
| chr9_81312837_81312988 | 0.08 |
PSAT1 |
phosphoserine aminotransferase 1 |
400853 |
0.01 |
| chr6_48103867_48104018 | 0.08 |
PTCHD4 |
patched domain containing 4 |
67517 |
0.14 |
| chr15_71571317_71571468 | 0.08 |
RP11-592N21.2 |
|
63271 |
0.13 |
| chr6_87836746_87836897 | 0.08 |
ZNF292 |
zinc finger protein 292 |
25730 |
0.15 |
| chr7_30405754_30405905 | 0.08 |
GS1-114I9.1 |
|
58459 |
0.11 |
| chr6_88407042_88408017 | 0.08 |
AKIRIN2 |
akirin 2 |
4398 |
0.23 |
| chr22_31985004_31985155 | 0.08 |
SFI1 |
Sfi1 homolog, spindle assembly associated (yeast) |
5187 |
0.21 |
| chr3_119631372_119631523 | 0.07 |
NR1I2 |
nuclear receptor subfamily 1, group I, member 2 |
129890 |
0.05 |
| chr15_52788876_52789027 | 0.07 |
MYO5A |
myosin VA (heavy chain 12, myoxin) |
32087 |
0.18 |
| chr9_7912412_7912563 | 0.07 |
TMEM261 |
transmembrane protein 261 |
112420 |
0.08 |
| chr11_128659981_128660132 | 0.07 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
25371 |
0.18 |
| chr6_141057887_141058038 | 0.07 |
ENSG00000264390 |
. |
53011 |
0.17 |
| chr6_151384328_151384479 | 0.07 |
RP1-292B18.3 |
|
7210 |
0.2 |
| chr4_54948926_54949077 | 0.07 |
GSX2 |
GS homeobox 2 |
17197 |
0.14 |
| chr7_139427311_139427656 | 0.07 |
HIPK2 |
homeodomain interacting protein kinase 2 |
5642 |
0.28 |
| chr19_2042508_2043083 | 0.07 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
163 |
0.91 |
| chr1_225942912_225943116 | 0.07 |
ENSG00000223306 |
. |
14037 |
0.14 |
| chr6_150949013_150949164 | 0.07 |
RP11-136K14.2 |
|
3275 |
0.27 |
| chr10_86095698_86095849 | 0.07 |
CCSER2 |
coiled-coil serine-rich protein 2 |
7354 |
0.26 |
| chr8_134089052_134089395 | 0.07 |
SLA |
Src-like-adaptor |
16620 |
0.22 |
| chr17_13766234_13766385 | 0.07 |
ENSG00000236088 |
. |
29847 |
0.25 |
| chr8_4852979_4853130 | 0.07 |
CSMD1 |
CUB and Sushi multiple domains 1 |
560 |
0.86 |
| chr6_84228850_84229001 | 0.07 |
PRSS35 |
protease, serine, 35 |
6668 |
0.28 |
| chr1_56839712_56839863 | 0.07 |
ENSG00000223307 |
. |
3303 |
0.39 |
| chr21_36879615_36879766 | 0.07 |
ENSG00000211590 |
. |
213323 |
0.02 |
| chr2_55346845_55347305 | 0.07 |
RTN4 |
reticulon 4 |
7318 |
0.2 |
| chrX_16051396_16051547 | 0.07 |
GRPR |
gastrin-releasing peptide receptor |
90208 |
0.08 |
| chr7_95239146_95239504 | 0.07 |
AC002451.3 |
|
217 |
0.94 |
| chr12_88718394_88718545 | 0.07 |
ENSG00000199245 |
. |
105750 |
0.07 |
| chr3_29294214_29294365 | 0.06 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
28184 |
0.21 |
| chr3_128958402_128958654 | 0.06 |
COPG1 |
coatomer protein complex, subunit gamma 1 |
9921 |
0.15 |
| chr3_69840030_69840181 | 0.06 |
ENSG00000241665 |
. |
21177 |
0.22 |
| chr19_13023555_13024013 | 0.06 |
SYCE2 |
synaptonemal complex central element protein 2 |
6306 |
0.06 |
| chr10_101753173_101753324 | 0.06 |
DNMBP |
dynamin binding protein |
16428 |
0.18 |
| chr5_9016573_9016856 | 0.06 |
CTD-2215L10.1 |
|
14828 |
0.25 |
| chr6_133281352_133281503 | 0.06 |
ENSG00000200534 |
. |
143069 |
0.04 |
| chr5_111498551_111498743 | 0.06 |
ENSG00000238363 |
. |
1465 |
0.41 |
| chr10_35006283_35006434 | 0.06 |
PARD3 |
par-3 family cell polarity regulator |
97891 |
0.08 |
| chr16_53290719_53290870 | 0.06 |
ENSG00000212582 |
. |
5959 |
0.22 |
| chr18_12409679_12409882 | 0.06 |
SLMO1 |
slowmo homolog 1 (Drosophila) |
1836 |
0.3 |
| chr2_201270022_201270173 | 0.06 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
12297 |
0.18 |
| chr7_100798830_100799198 | 0.06 |
AP1S1 |
adaptor-related protein complex 1, sigma 1 subunit |
1208 |
0.28 |
| chr11_79125207_79125358 | 0.06 |
ENSG00000266570 |
. |
7988 |
0.21 |
| chr12_13500086_13500237 | 0.06 |
C12orf36 |
chromosome 12 open reading frame 36 |
29203 |
0.21 |
| chr7_134584974_134585125 | 0.06 |
CALD1 |
caldesmon 1 |
8688 |
0.28 |
| chr12_121727254_121727405 | 0.06 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
7160 |
0.21 |
| chr1_94371593_94371744 | 0.06 |
GCLM |
glutamate-cysteine ligase, modifier subunit |
3298 |
0.25 |
| chr10_121572117_121572268 | 0.06 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
6036 |
0.23 |
| chr8_130334106_130334257 | 0.06 |
ENSG00000222755 |
. |
30952 |
0.22 |
| chr3_131104607_131104908 | 0.06 |
NUDT16 |
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
4128 |
0.2 |
| chr7_127421938_127422089 | 0.06 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
105952 |
0.06 |
| chr14_32842058_32842209 | 0.05 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
43527 |
0.15 |
| chr12_109118549_109119150 | 0.05 |
CORO1C |
coronin, actin binding protein, 1C |
5478 |
0.17 |
| chr14_68633954_68634105 | 0.05 |
ENSG00000244677 |
. |
17404 |
0.22 |
| chr4_129375310_129375461 | 0.05 |
PGRMC2 |
progesterone receptor membrane component 2 |
165401 |
0.04 |
| chr11_10064252_10064435 | 0.05 |
SBF2 |
SET binding factor 2 |
49692 |
0.16 |
| chr1_234641581_234641923 | 0.05 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
26903 |
0.19 |
| chr12_69743933_69744084 | 0.05 |
LYZ |
lysozyme |
1842 |
0.3 |
| chr11_114034689_114034840 | 0.05 |
ENSG00000221112 |
. |
77113 |
0.09 |
| chr18_56237913_56238064 | 0.05 |
RP11-126O1.2 |
|
13686 |
0.16 |
| chr6_74498861_74499012 | 0.05 |
RP11-553A21.3 |
|
93082 |
0.08 |
| chr6_15389566_15390143 | 0.05 |
JARID2 |
jumonji, AT rich interactive domain 2 |
11235 |
0.25 |
| chr1_12674943_12675094 | 0.05 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
2719 |
0.25 |
| chr19_45981127_45981279 | 0.05 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
883 |
0.43 |
| chr8_52863208_52863359 | 0.05 |
RP11-110G21.1 |
|
50989 |
0.13 |
| chr18_10269485_10269636 | 0.05 |
ENSG00000239031 |
. |
120437 |
0.06 |
| chr17_40388027_40388178 | 0.05 |
RP11-358B23.5 |
|
32503 |
0.08 |
| chr10_116474466_116474617 | 0.05 |
ABLIM1 |
actin binding LIM protein 1 |
30127 |
0.21 |
| chr1_113166689_113166984 | 0.05 |
ST7L |
suppression of tumorigenicity 7 like |
4796 |
0.14 |
| chr9_95355824_95355975 | 0.05 |
CENPP |
centromere protein P |
6640 |
0.21 |
| chr2_36364447_36364598 | 0.04 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
218547 |
0.02 |
| chr17_8870824_8871109 | 0.04 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
1942 |
0.4 |
| chr5_149185316_149185608 | 0.04 |
PPARGC1B |
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
27116 |
0.17 |
| chr14_24611105_24611656 | 0.04 |
EMC9 |
ER membrane protein complex subunit 9 |
583 |
0.43 |
| chr10_114673586_114673994 | 0.04 |
RP11-57H14.3 |
|
25296 |
0.18 |
| chr11_36622535_36622686 | 0.04 |
RAG2 |
recombination activating gene 2 |
2824 |
0.27 |
| chr16_86623972_86624123 | 0.04 |
FOXL1 |
forkhead box L1 |
11932 |
0.17 |
| chr21_30691484_30691635 | 0.04 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
7891 |
0.19 |
| chr8_131269264_131269415 | 0.04 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
10051 |
0.29 |
| chr1_81659170_81659383 | 0.04 |
ENSG00000223026 |
. |
58198 |
0.16 |
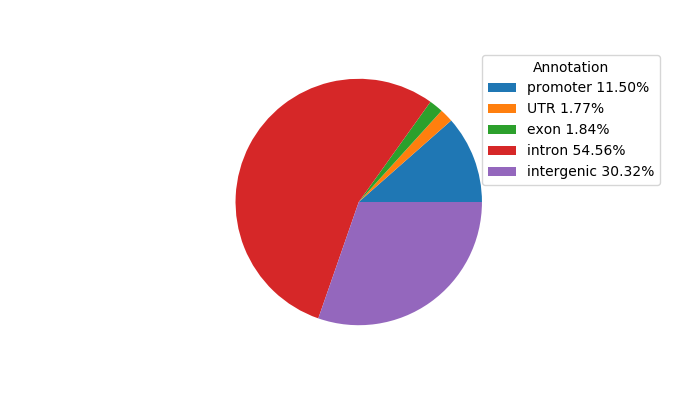
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0090568 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |