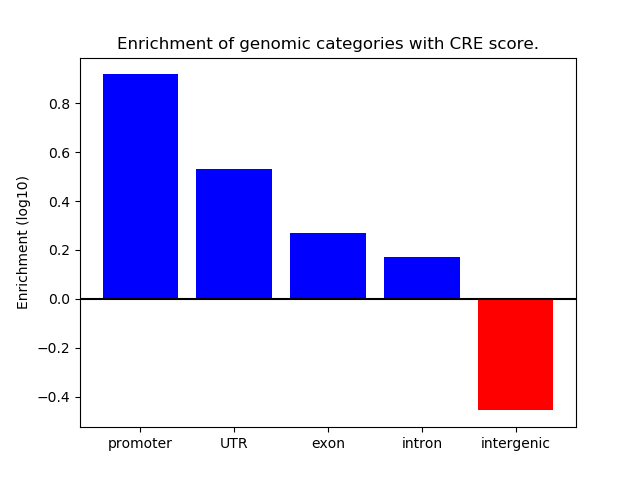
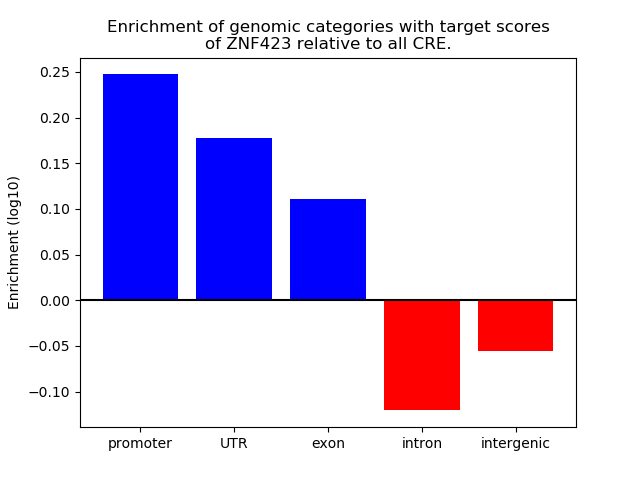
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
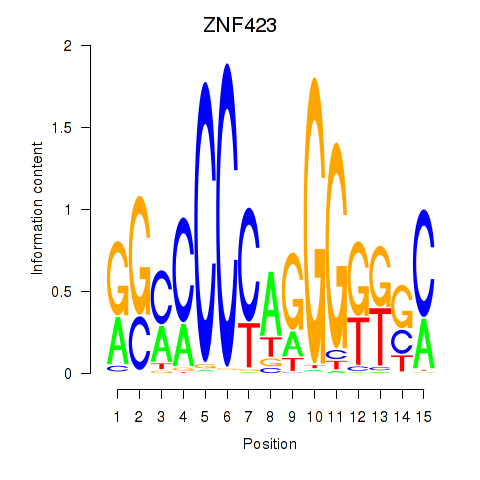
Results for ZNF423
Z-value: 1.27
Transcription factors associated with ZNF423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF423
|
ENSG00000102935.7 | zinc finger protein 423 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
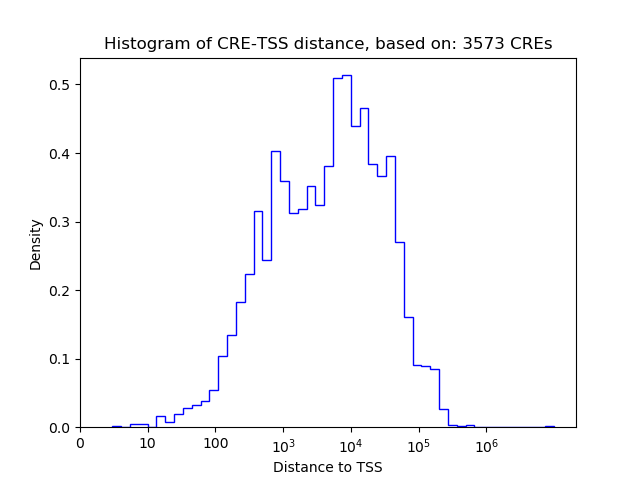
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_49890174_49890551 | ZNF423 | 316 | 0.923061 | 0.86 | 3.0e-03 | Click! |
| chr16_49891743_49892055 | ZNF423 | 69 | 0.981476 | 0.75 | 2.0e-02 | Click! |
| chr16_49892831_49893014 | ZNF423 | 1092 | 0.601079 | 0.73 | 2.4e-02 | Click! |
| chr16_49683940_49684091 | ZNF423 | 9252 | 0.251366 | -0.73 | 2.5e-02 | Click! |
| chr16_49892506_49892687 | ZNF423 | 766 | 0.728454 | 0.71 | 3.1e-02 | Click! |
Activity of the ZNF423 motif across conditions
Conditions sorted by the z-value of the ZNF423 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
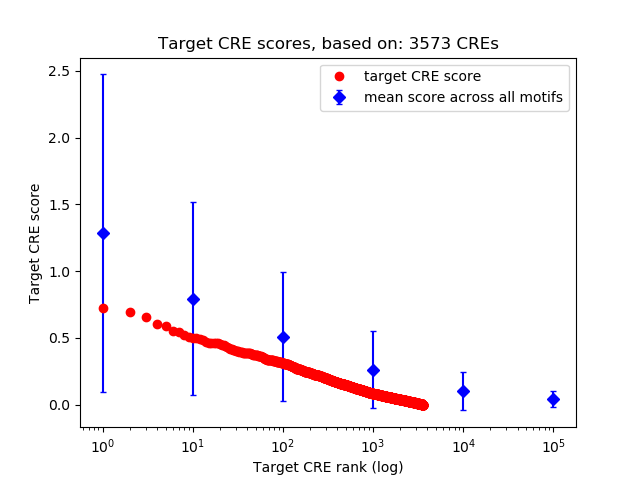
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
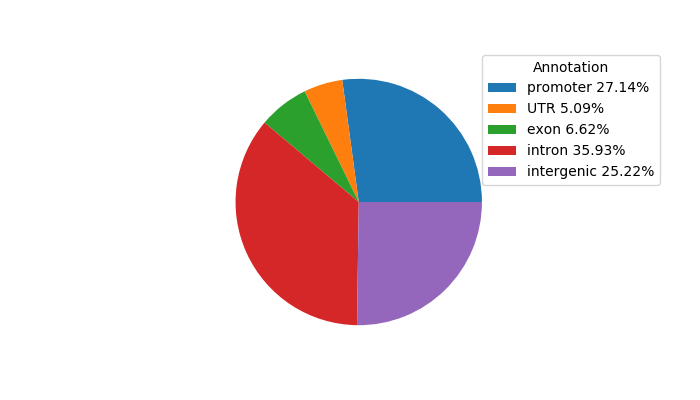
| chr1_25886127_25886448 | 0.73 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
16216 |
0.19 |
| chr16_68109101_68109477 | 0.70 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
9958 |
0.1 |
| chr16_89521002_89521377 | 0.65 |
ENSG00000252887 |
. |
11463 |
0.12 |
| chr19_19737216_19737375 | 0.60 |
LPAR2 |
lysophosphatidic acid receptor 2 |
1314 |
0.3 |
| chr16_85481767_85482285 | 0.59 |
ENSG00000264203 |
. |
6928 |
0.27 |
| chr1_117303582_117303773 | 0.55 |
CD2 |
CD2 molecule |
6588 |
0.23 |
| chr5_133440168_133440408 | 0.55 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
10114 |
0.23 |
| chr17_45277609_45277834 | 0.52 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
8680 |
0.12 |
| chr16_31477943_31478110 | 0.51 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
4880 |
0.1 |
| chr9_130473685_130473878 | 0.50 |
C9orf117 |
chromosome 9 open reading frame 117 |
388 |
0.73 |
| chr15_44969737_44969888 | 0.50 |
PATL2 |
protein associated with topoisomerase II homolog 2 (yeast) |
726 |
0.6 |
| chr14_99673860_99674011 | 0.49 |
AL162151.4 |
|
49182 |
0.14 |
| chr6_20206050_20206262 | 0.49 |
RP11-239H6.2 |
|
6162 |
0.24 |
| chr11_75220135_75220483 | 0.47 |
RP11-939C17.4 |
|
873 |
0.54 |
| chr2_28647839_28647990 | 0.46 |
RP11-373D23.2 |
|
28232 |
0.14 |
| chr14_94422859_94423119 | 0.46 |
ASB2 |
ankyrin repeat and SOCS box containing 2 |
778 |
0.58 |
| chr2_173090506_173090657 | 0.46 |
ENSG00000238572 |
. |
69733 |
0.11 |
| chr4_153033286_153033437 | 0.46 |
ENSG00000266244 |
. |
116259 |
0.06 |
| chr17_75842366_75842622 | 0.46 |
FLJ45079 |
|
36165 |
0.19 |
| chr20_61639034_61639185 | 0.46 |
BHLHE23 |
basic helix-loop-helix family, member e23 |
722 |
0.67 |
| chr22_19585555_19585986 | 0.44 |
CLDN5 |
claudin 5 |
70702 |
0.08 |
| chr1_9776346_9776497 | 0.44 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
1706 |
0.34 |
| chr1_25345075_25345361 | 0.44 |
ENSG00000264371 |
. |
4776 |
0.25 |
| chr21_43579719_43579870 | 0.44 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
40005 |
0.13 |
| chr16_89762519_89762771 | 0.42 |
SPATA2L |
spermatogenesis associated 2-like |
5403 |
0.1 |
| chr3_51718621_51718776 | 0.42 |
ENSG00000201595 |
. |
9900 |
0.15 |
| chr22_46769711_46769909 | 0.42 |
TRMU |
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
37991 |
0.15 |
| chr2_235333773_235334004 | 0.41 |
ARL4C |
ADP-ribosylation factor-like 4C |
71356 |
0.13 |
| chr14_91832903_91833121 | 0.41 |
ENSG00000265856 |
. |
32955 |
0.17 |
| chr14_105662119_105662372 | 0.40 |
NUDT14 |
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
14605 |
0.15 |
| chr19_54609905_54610056 | 0.40 |
NDUFA3 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
708 |
0.42 |
| chr3_169583062_169583383 | 0.40 |
LRRC31 |
leucine rich repeat containing 31 |
4399 |
0.16 |
| chr17_37832739_37832926 | 0.40 |
PNMT |
phenylethanolamine N-methyltransferase |
8116 |
0.1 |
| chr22_31289621_31289772 | 0.39 |
OSBP2 |
oxysterol binding protein 2 |
468 |
0.8 |
| chr5_9544937_9545108 | 0.39 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
1165 |
0.47 |
| chr4_2792015_2792228 | 0.39 |
SH3BP2 |
SH3-domain binding protein 2 |
2629 |
0.29 |
| chr5_67579614_67579808 | 0.39 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
3576 |
0.36 |
| chr1_777188_777339 | 0.39 |
RP11-206L10.8 |
|
31722 |
0.11 |
| chr1_200856866_200857017 | 0.39 |
C1orf106 |
chromosome 1 open reading frame 106 |
3235 |
0.23 |
| chr19_54378263_54378435 | 0.39 |
AC008440.5 |
|
954 |
0.29 |
| chr4_2836679_2837097 | 0.39 |
ADD1 |
adducin 1 (alpha) |
8696 |
0.19 |
| chr11_68597072_68597223 | 0.39 |
CPT1A |
carnitine palmitoyltransferase 1A (liver) |
10005 |
0.2 |
| chr15_69101431_69101582 | 0.38 |
ENSG00000265195 |
. |
7242 |
0.23 |
| chr1_117453586_117453737 | 0.38 |
PTGFRN |
prostaglandin F2 receptor inhibitor |
982 |
0.54 |
| chr2_70336914_70337228 | 0.38 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
21093 |
0.14 |
| chr12_108963200_108963590 | 0.38 |
ISCU |
iron-sulfur cluster assembly enzyme |
7009 |
0.15 |
| chr9_134127671_134127998 | 0.37 |
FAM78A |
family with sequence similarity 78, member A |
18046 |
0.15 |
| chr19_30160205_30160356 | 0.37 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
2499 |
0.33 |
| chr1_92931900_92932072 | 0.37 |
GFI1 |
growth factor independent 1 transcription repressor |
17525 |
0.23 |
| chr1_203291434_203291646 | 0.37 |
ENSG00000202300 |
. |
3313 |
0.22 |
| chr8_134508972_134509123 | 0.37 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
2557 |
0.42 |
| chr6_167505175_167505326 | 0.37 |
CCR6 |
chemokine (C-C motif) receptor 6 |
20045 |
0.18 |
| chr10_30985837_30986131 | 0.37 |
SVILP1 |
supervillin pseudogene 1 |
1134 |
0.61 |
| chr2_46118370_46118521 | 0.37 |
PRKCE |
protein kinase C, epsilon |
109596 |
0.07 |
| chr11_62408838_62409025 | 0.37 |
GANAB |
glucosidase, alpha; neutral AB |
5150 |
0.06 |
| chr6_15318009_15318227 | 0.37 |
ENSG00000201367 |
. |
2967 |
0.27 |
| chr1_42271688_42271993 | 0.36 |
ENSG00000264896 |
. |
47028 |
0.18 |
| chr20_62475435_62475724 | 0.36 |
AL158091.1 |
Protein LOC100509861 |
306 |
0.78 |
| chr12_123571415_123571566 | 0.36 |
PITPNM2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
5610 |
0.2 |
| chr2_127976471_127976630 | 0.35 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
1104 |
0.58 |
| chr21_46900861_46901012 | 0.35 |
COL18A1 |
collagen, type XVIII, alpha 1 |
9253 |
0.2 |
| chr10_12589202_12589377 | 0.35 |
ENSG00000263584 |
. |
31463 |
0.2 |
| chr1_1105743_1105894 | 0.35 |
ENSG00000198976 |
. |
1433 |
0.19 |
| chr17_45780207_45780358 | 0.34 |
TBKBP1 |
TBK1 binding protein 1 |
7652 |
0.14 |
| chr1_234306113_234306370 | 0.34 |
SLC35F3 |
solute carrier family 35, member F3 |
43776 |
0.14 |
| chrX_128902828_128903127 | 0.34 |
SASH3 |
SAM and SH3 domain containing 3 |
10983 |
0.19 |
| chr9_92445291_92445531 | 0.34 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
225458 |
0.02 |
| chr1_42106059_42106288 | 0.34 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
60499 |
0.13 |
| chr9_92111235_92111454 | 0.34 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
1003 |
0.5 |
| chr1_167656783_167656934 | 0.34 |
MPZL1 |
myelin protein zero-like 1 |
34329 |
0.16 |
| chr4_1617642_1617793 | 0.33 |
FAM53A |
family with sequence similarity 53, member A |
39418 |
0.14 |
| chr5_169687872_169688139 | 0.33 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
6295 |
0.24 |
| chr16_88770397_88770548 | 0.33 |
RNF166 |
ring finger protein 166 |
447 |
0.59 |
| chr7_20370436_20370587 | 0.33 |
ITGB8 |
integrin, beta 8 |
186 |
0.88 |
| chr1_54844897_54845115 | 0.33 |
SSBP3 |
single stranded DNA binding protein 3 |
26171 |
0.19 |
| chr9_107877201_107877485 | 0.33 |
ENSG00000201583 |
. |
18306 |
0.27 |
| chr11_65385759_65385910 | 0.33 |
PCNXL3 |
pecanex-like 3 (Drosophila) |
2590 |
0.11 |
| chr1_112020736_112021030 | 0.33 |
C1orf162 |
chromosome 1 open reading frame 162 |
4392 |
0.12 |
| chr9_134458873_134459024 | 0.33 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
38277 |
0.13 |
| chr21_45334011_45334235 | 0.33 |
AGPAT3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
2030 |
0.3 |
| chr17_72459036_72459537 | 0.33 |
CD300A |
CD300a molecule |
3269 |
0.19 |
| chr11_117821389_117821783 | 0.33 |
TMPRSS13 |
transmembrane protease, serine 13 |
21412 |
0.15 |
| chr6_158993933_158994198 | 0.33 |
TMEM181 |
transmembrane protein 181 |
36597 |
0.15 |
| chr7_138777169_138777799 | 0.32 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
13471 |
0.2 |
| chr9_95825508_95825700 | 0.32 |
SUSD3 |
sushi domain containing 3 |
4543 |
0.2 |
| chr11_1606465_1606821 | 0.32 |
KRTAP5-1 |
keratin associated protein 5-1 |
130 |
0.93 |
| chr3_187767301_187767602 | 0.32 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
103621 |
0.07 |
| chr10_29776432_29776729 | 0.32 |
SVIL |
supervillin |
8867 |
0.24 |
| chr22_30693041_30693254 | 0.32 |
RP1-130H16.18 |
Uncharacterized protein |
2324 |
0.16 |
| chr14_98459359_98459510 | 0.32 |
C14orf64 |
chromosome 14 open reading frame 64 |
14973 |
0.31 |
| chr15_70825685_70825836 | 0.32 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
168860 |
0.04 |
| chr1_167570557_167570959 | 0.32 |
RCSD1 |
RCSD domain containing 1 |
28572 |
0.15 |
| chr8_128994096_128994247 | 0.32 |
ENSG00000207110 |
. |
17206 |
0.16 |
| chr20_1677648_1677799 | 0.32 |
ENSG00000242348 |
. |
30717 |
0.14 |
| chr6_125666901_125667052 | 0.32 |
RP11-735G4.1 |
|
28494 |
0.21 |
| chr7_102072492_102072712 | 0.32 |
ORAI2 |
ORAI calcium release-activated calcium modulator 2 |
951 |
0.34 |
| chr2_99077021_99077380 | 0.32 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
15787 |
0.23 |
| chr5_154193336_154193487 | 0.31 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
9630 |
0.16 |
| chr21_19152626_19152837 | 0.31 |
AL109761.5 |
|
13074 |
0.23 |
| chr8_97165985_97166258 | 0.31 |
GDF6 |
growth differentiation factor 6 |
6899 |
0.26 |
| chr17_73234704_73234855 | 0.31 |
GGA3 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
287 |
0.81 |
| chr9_36143975_36144155 | 0.31 |
GLIPR2 |
GLI pathogenesis-related 2 |
7323 |
0.19 |
| chr1_2426510_2426661 | 0.31 |
RP3-395M20.3 |
|
667 |
0.51 |
| chr6_24866477_24866656 | 0.31 |
FAM65B |
family with sequence similarity 65, member B |
10975 |
0.19 |
| chr1_204450638_204450997 | 0.31 |
PIK3C2B |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
8629 |
0.19 |
| chr20_39774881_39775244 | 0.31 |
RP1-1J6.2 |
|
8419 |
0.2 |
| chr17_76115212_76115363 | 0.30 |
TMC6 |
transmembrane channel-like 6 |
3542 |
0.15 |
| chr22_29215628_29215779 | 0.30 |
CTA-292E10.6 |
|
1384 |
0.35 |
| chr1_118195260_118195411 | 0.30 |
ENSG00000212266 |
. |
36029 |
0.17 |
| chr1_183440737_183440894 | 0.30 |
SMG7 |
SMG7 nonsense mediated mRNA decay factor |
691 |
0.73 |
| chr20_47239118_47239269 | 0.30 |
ENSG00000238452 |
. |
116638 |
0.06 |
| chr7_4792903_4793054 | 0.30 |
FOXK1 |
forkhead box K1 |
12488 |
0.16 |
| chr19_42398087_42398358 | 0.30 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
143 |
0.93 |
| chr15_29382386_29382685 | 0.30 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
46008 |
0.17 |
| chr10_135076415_135076765 | 0.30 |
ADAM8 |
ADAM metallopeptidase domain 8 |
13764 |
0.09 |
| chr20_55975873_55976024 | 0.30 |
RP4-800J21.3 |
|
7830 |
0.16 |
| chr17_45782038_45782291 | 0.30 |
TBKBP1 |
TBK1 binding protein 1 |
9534 |
0.13 |
| chr18_21200887_21201038 | 0.30 |
ANKRD29 |
ankyrin repeat domain 29 |
28524 |
0.15 |
| chr14_101109543_101109694 | 0.30 |
CTD-2644I21.1 |
|
53955 |
0.07 |
| chr9_127120006_127120171 | 0.30 |
ENSG00000264237 |
. |
7278 |
0.21 |
| chr7_1104722_1104898 | 0.29 |
RP11-449P15.1 |
|
5913 |
0.1 |
| chr7_49814485_49814636 | 0.29 |
VWC2 |
von Willebrand factor C domain containing 2 |
1303 |
0.63 |
| chr16_67795135_67795286 | 0.29 |
RP11-167P11.2 |
|
21385 |
0.1 |
| chr21_46333137_46333552 | 0.29 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
854 |
0.44 |
| chr5_176802584_176802735 | 0.29 |
SLC34A1 |
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
3577 |
0.12 |
| chr14_91849387_91849607 | 0.29 |
CCDC88C |
coiled-coil domain containing 88C |
34193 |
0.17 |
| chr6_139494692_139494843 | 0.29 |
HECA |
headcase homolog (Drosophila) |
38518 |
0.17 |
| chr15_81588541_81588692 | 0.28 |
IL16 |
interleukin 16 |
638 |
0.74 |
| chr22_37812190_37812341 | 0.28 |
RP1-63G5.5 |
|
11215 |
0.14 |
| chr5_169706251_169706453 | 0.28 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
12021 |
0.22 |
| chr11_63305751_63305902 | 0.28 |
RARRES3 |
retinoic acid receptor responder (tazarotene induced) 3 |
42 |
0.97 |
| chr6_33663289_33663440 | 0.28 |
SBP1 |
SBP1; Uncharacterized protein |
110 |
0.95 |
| chr20_50143060_50143387 | 0.28 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
16035 |
0.26 |
| chr1_12245359_12245510 | 0.28 |
ENSG00000263676 |
. |
6336 |
0.17 |
| chr19_50923202_50923353 | 0.28 |
SPIB |
Spi-B transcription factor (Spi-1/PU.1 related) |
1062 |
0.3 |
| chr11_6742431_6742729 | 0.28 |
GVINP1 |
GTPase, very large interferon inducible pseudogene 1 |
531 |
0.69 |
| chr14_100850226_100850465 | 0.28 |
WDR25 |
WD repeat domain 25 |
2008 |
0.19 |
| chr7_142045834_142045985 | 0.28 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
56300 |
0.13 |
| chr11_74983514_74983728 | 0.27 |
ARRB1 |
arrestin, beta 1 |
6122 |
0.14 |
| chr4_1797283_1797487 | 0.27 |
FGFR3 |
fibroblast growth factor receptor 3 |
1762 |
0.33 |
| chr20_62104787_62104938 | 0.27 |
KCNQ2 |
potassium voltage-gated channel, KQT-like subfamily, member 2 |
869 |
0.46 |
| chr20_37435383_37435534 | 0.27 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
1091 |
0.52 |
| chr2_95716411_95716700 | 0.27 |
AC103563.9 |
|
2366 |
0.25 |
| chr20_62488359_62488510 | 0.27 |
ABHD16B |
abhydrolase domain containing 16B |
4132 |
0.09 |
| chr17_1199306_1199457 | 0.27 |
TUSC5 |
tumor suppressor candidate 5 |
16424 |
0.17 |
| chr2_99449405_99449556 | 0.27 |
ENSG00000238830 |
. |
7719 |
0.22 |
| chr15_70545445_70545596 | 0.27 |
ENSG00000200216 |
. |
59945 |
0.15 |
| chr11_67177814_67178291 | 0.27 |
CARNS1 |
carnosine synthase 1 |
5097 |
0.07 |
| chr3_18684786_18684937 | 0.27 |
ENSG00000228956 |
. |
50518 |
0.19 |
| chr2_238813522_238813924 | 0.27 |
ENSG00000263723 |
. |
35176 |
0.15 |
| chr1_154932014_154932227 | 0.27 |
PYGO2 |
pygopus family PHD finger 2 |
2104 |
0.12 |
| chr4_2838265_2838548 | 0.27 |
ADD1 |
adducin 1 (alpha) |
7178 |
0.2 |
| chr11_67201895_67202105 | 0.27 |
AP003419.16 |
|
870 |
0.3 |
| chr17_77904427_77904652 | 0.26 |
TBC1D16 |
TBC1 domain family, member 16 |
20088 |
0.18 |
| chr13_53277825_53277976 | 0.26 |
LECT1 |
leukocyte cell derived chemotaxin 1 |
35587 |
0.16 |
| chr17_72474398_72474597 | 0.26 |
CD300A |
CD300a molecule |
11496 |
0.14 |
| chr2_74228872_74229028 | 0.26 |
TET3 |
tet methylcytosine dioxygenase 3 |
890 |
0.55 |
| chr13_111948297_111948901 | 0.26 |
TEX29 |
testis expressed 29 |
24416 |
0.22 |
| chr3_15319690_15320104 | 0.26 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
2345 |
0.23 |
| chr11_441488_441639 | 0.26 |
ANO9 |
anoctamin 9 |
448 |
0.56 |
| chr22_42592642_42592793 | 0.26 |
TCF20 |
transcription factor 20 (AR1) |
13497 |
0.16 |
| chr1_207095862_207096179 | 0.26 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
521 |
0.73 |
| chr19_3024526_3024677 | 0.26 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
424 |
0.77 |
| chr16_88535878_88536112 | 0.26 |
ENSG00000263456 |
. |
669 |
0.68 |
| chr17_43310695_43311082 | 0.26 |
FMNL1 |
formin-like 1 |
7554 |
0.1 |
| chr19_4088417_4088592 | 0.26 |
MAP2K2 |
mitogen-activated protein kinase kinase 2 |
12541 |
0.11 |
| chr17_73150023_73150174 | 0.25 |
HN1 |
hematological and neurological expressed 1 |
137 |
0.91 |
| chr10_82339230_82339422 | 0.25 |
SH2D4B |
SH2 domain containing 4B |
38751 |
0.17 |
| chr2_106429372_106429523 | 0.25 |
NCK2 |
NCK adaptor protein 2 |
3568 |
0.32 |
| chr19_50064241_50064618 | 0.25 |
NOSIP |
nitric oxide synthase interacting protein |
481 |
0.57 |
| chr9_92063731_92064015 | 0.25 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
6995 |
0.26 |
| chr11_128555736_128555887 | 0.25 |
RP11-744N12.3 |
|
512 |
0.52 |
| chr9_136351101_136351399 | 0.25 |
SLC2A6 |
solute carrier family 2 (facilitated glucose transporter), member 6 |
6991 |
0.12 |
| chr12_12418765_12419238 | 0.25 |
LRP6 |
low density lipoprotein receptor-related protein 6 |
702 |
0.68 |
| chr22_44890267_44890418 | 0.25 |
LDOC1L |
leucine zipper, down-regulated in cancer 1-like |
3836 |
0.34 |
| chr4_54959914_54960069 | 0.25 |
GSX2 |
GS homeobox 2 |
6207 |
0.17 |
| chr11_2860656_2860807 | 0.25 |
KCNQ1-AS1 |
KCNQ1 antisense RNA 1 |
22067 |
0.13 |
| chr19_4064720_4064871 | 0.25 |
ZBTB7A |
zinc finger and BTB domain containing 7A |
935 |
0.4 |
| chr12_133100245_133100396 | 0.25 |
FBRSL1 |
fibrosin-like 1 |
33163 |
0.16 |
| chr13_99176253_99176430 | 0.25 |
STK24 |
serine/threonine kinase 24 |
2089 |
0.37 |
| chr3_13057227_13057378 | 0.25 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
28766 |
0.23 |
| chr2_27530014_27530299 | 0.25 |
UCN |
urocortin |
1157 |
0.27 |
| chr7_75367789_75368030 | 0.25 |
HIP1 |
huntingtin interacting protein 1 |
350 |
0.89 |
| chr1_214460978_214461129 | 0.25 |
SMYD2 |
SET and MYND domain containing 2 |
6477 |
0.3 |
| chr22_50754075_50754226 | 0.25 |
XX-C283C717.1 |
|
1090 |
0.31 |
| chr19_12550187_12550350 | 0.24 |
CTD-3105H18.16 |
Uncharacterized protein |
1628 |
0.22 |
| chrX_46488224_46488448 | 0.24 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
55117 |
0.14 |
| chr17_8533635_8533853 | 0.24 |
MYH10 |
myosin, heavy chain 10, non-muscle |
291 |
0.92 |
| chr12_104350569_104350720 | 0.24 |
C12orf73 |
chromosome 12 open reading frame 73 |
281 |
0.87 |
| chr17_48988068_48988440 | 0.24 |
TOB1 |
transducer of ERBB2, 1 |
42915 |
0.11 |
| chr1_9776061_9776212 | 0.24 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
1991 |
0.31 |
| chr21_46741518_46741669 | 0.24 |
ENSG00000215447 |
. |
33626 |
0.13 |
| chr3_13277082_13277233 | 0.24 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
162540 |
0.04 |
| chr20_61424831_61424989 | 0.24 |
MRGBP |
MRG/MORF4L binding protein |
2895 |
0.16 |
| chr1_151793784_151793935 | 0.24 |
RORC |
RAR-related orphan receptor C |
4708 |
0.09 |
| chr22_27067872_27068143 | 0.24 |
CRYBA4 |
crystallin, beta A4 |
50079 |
0.13 |
| chr19_7741444_7741683 | 0.24 |
C19orf59 |
chromosome 19 open reading frame 59 |
49 |
0.93 |
| chr7_151078058_151078311 | 0.24 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
12268 |
0.13 |
| chr13_114875590_114875798 | 0.24 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
1399 |
0.48 |
| chr2_242800699_242800986 | 0.24 |
PDCD1 |
programmed cell death 1 |
218 |
0.89 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.1 | 0.2 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.1 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.1 | 0.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0051446 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.2 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.2 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.3 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.2 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.0 | 0.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0014808 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.2 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.2 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.2 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.0 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0032769 | negative regulation of monooxygenase activity(GO:0032769) |
| 0.0 | 0.8 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.1 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.2 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0061054 | dermatome development(GO:0061054) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.2 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.0 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.1 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.0 | GO:0002724 | regulation of T cell cytokine production(GO:0002724) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:0032610 | interleukin-1 alpha production(GO:0032610) regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.0 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.0 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.7 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.3 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.0 | 0.1 | GO:0032967 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.1 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:0002209 | behavioral defense response(GO:0002209) |
| 0.0 | 0.1 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.0 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.0 | GO:0090185 | negative regulation of kidney development(GO:0090185) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.0 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.0 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) |
| 0.0 | 0.1 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0006007 | glucose catabolic process(GO:0006007) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.0 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.2 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.0 | GO:0002448 | mast cell mediated immunity(GO:0002448) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031083 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0030288 | outer membrane-bounded periplasmic space(GO:0030288) cell envelope(GO:0030313) periplasmic space(GO:0042597) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.2 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.0 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.0 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |