Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
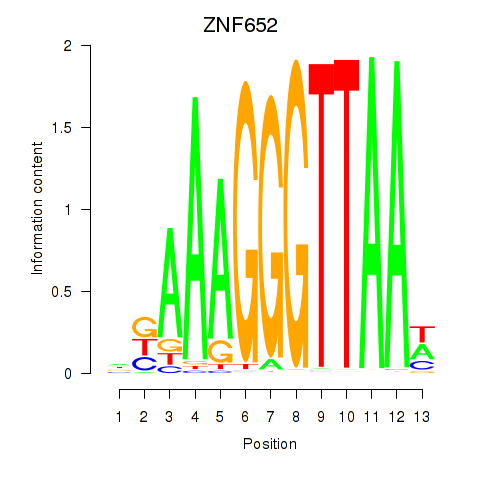
Results for ZNF652
Z-value: 1.21
Transcription factors associated with ZNF652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF652
|
ENSG00000198740.4 | zinc finger protein 652 |
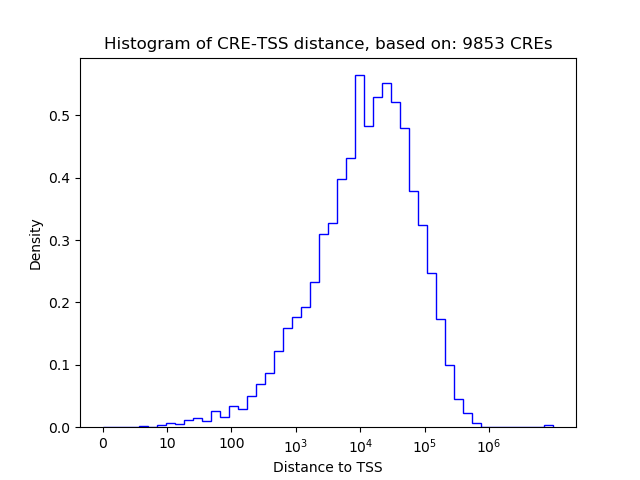
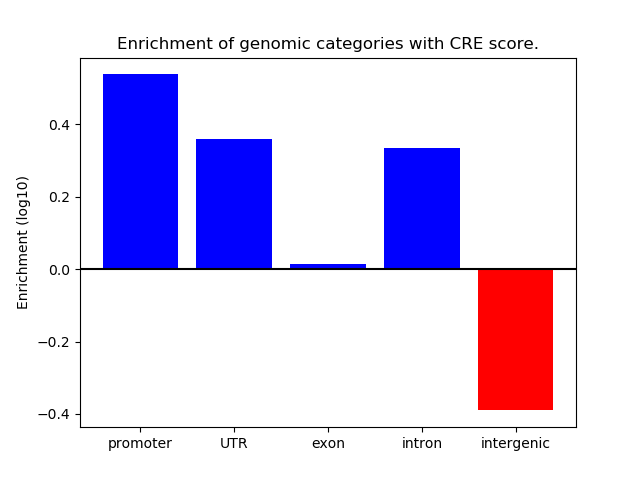
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_47437924_47438149 | ZNF652 | 1442 | 0.297279 | -0.49 | 1.8e-01 | Click! |
| chr17_47427033_47427184 | ZNF652 | 12370 | 0.121873 | 0.42 | 2.7e-01 | Click! |
| chr17_47422139_47422537 | ZNF652 | 17140 | 0.117289 | 0.26 | 4.9e-01 | Click! |
| chr17_47426040_47426191 | ZNF652 | 13363 | 0.120905 | 0.24 | 5.3e-01 | Click! |
| chr17_47438224_47438740 | ZNF652 | 996 | 0.421898 | -0.22 | 5.7e-01 | Click! |
Activity of the ZNF652 motif across conditions
Conditions sorted by the z-value of the ZNF652 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
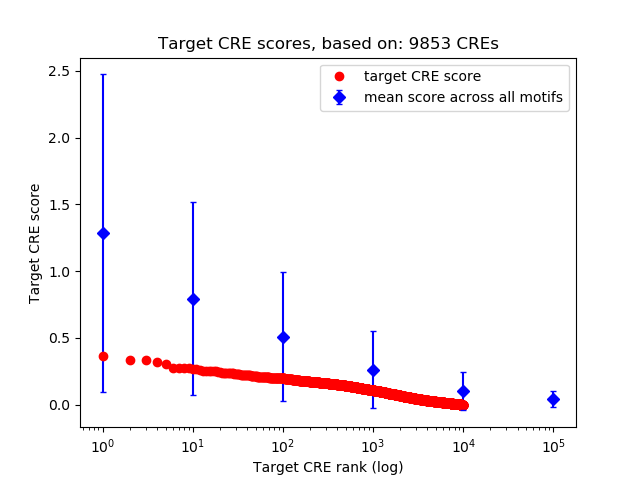
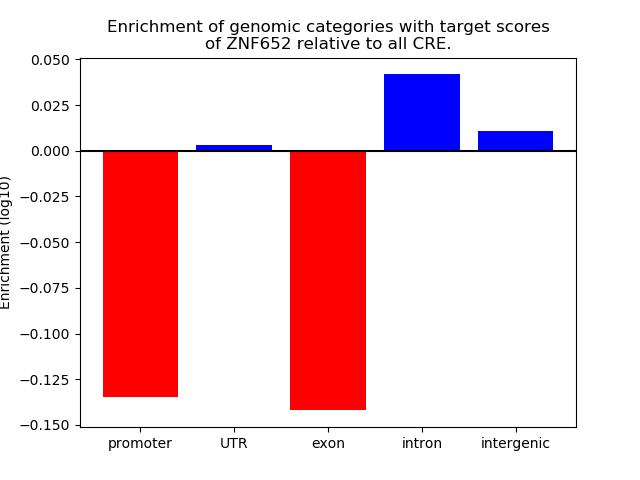
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_45167020_45167995 | 0.36 |
PRDM11 |
PR domain containing 11 |
719 |
0.77 |
| chr7_128467967_128468227 | 0.34 |
FLNC |
filamin C, gamma |
2334 |
0.21 |
| chr17_13443526_13443677 | 0.33 |
ENSG00000221698 |
. |
3362 |
0.28 |
| chr2_158161910_158162061 | 0.32 |
ENSG00000222404 |
. |
6315 |
0.23 |
| chr6_44018695_44019099 | 0.31 |
RP5-1120P11.1 |
|
23492 |
0.15 |
| chr6_121841417_121841568 | 0.28 |
ENSG00000201379 |
. |
22280 |
0.2 |
| chr1_162828153_162828448 | 0.28 |
C1orf110 |
chromosome 1 open reading frame 110 |
10251 |
0.24 |
| chr7_131311005_131311156 | 0.27 |
PODXL |
podocalyxin-like |
69704 |
0.13 |
| chr3_65980527_65980678 | 0.27 |
ENSG00000202071 |
. |
9013 |
0.2 |
| chr1_207072109_207072264 | 0.27 |
IL24 |
interleukin 24 |
1008 |
0.47 |
| chr9_84387097_84387248 | 0.27 |
ENSG00000263404 |
. |
33824 |
0.17 |
| chr9_694654_694862 | 0.26 |
RP11-130C19.3 |
|
9203 |
0.22 |
| chr2_32703898_32704049 | 0.26 |
BIRC6 |
baculoviral IAP repeat containing 6 |
45257 |
0.15 |
| chr9_129895370_129895521 | 0.25 |
ANGPTL2 |
angiopoietin-like 2 |
10283 |
0.23 |
| chr19_11086277_11086433 | 0.25 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
8473 |
0.14 |
| chr6_133228663_133228814 | 0.25 |
ENSG00000200534 |
. |
90380 |
0.06 |
| chr3_23855395_23855546 | 0.25 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
2521 |
0.28 |
| chr3_8503582_8503733 | 0.25 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
39627 |
0.16 |
| chr17_30518069_30518363 | 0.24 |
AC116407.2 |
Uncharacterized protein |
11331 |
0.14 |
| chr22_31226560_31226711 | 0.24 |
OSBP2 |
oxysterol binding protein 2 |
8132 |
0.21 |
| chr16_585344_585519 | 0.24 |
ENSG00000266124 |
. |
115 |
0.91 |
| chr3_8499065_8499317 | 0.24 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
44093 |
0.15 |
| chr1_94791983_94792377 | 0.24 |
ARHGAP29 |
Rho GTPase activating protein 29 |
88991 |
0.08 |
| chrX_46442028_46442386 | 0.24 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
8988 |
0.23 |
| chr3_67697418_67697833 | 0.24 |
SUCLG2 |
succinate-CoA ligase, GDP-forming, beta subunit |
7406 |
0.34 |
| chr5_77996016_77996172 | 0.24 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
51446 |
0.18 |
| chr19_15559479_15560330 | 0.23 |
ENSG00000269782 |
. |
455 |
0.47 |
| chr7_102802458_102802750 | 0.23 |
NAPEPLD |
N-acyl phosphatidylethanolamine phospholipase D |
12597 |
0.21 |
| chr12_46886917_46887068 | 0.23 |
SLC38A2 |
solute carrier family 38, member 2 |
120342 |
0.06 |
| chr1_109045628_109045779 | 0.23 |
NBPF6 |
neuroblastoma breakpoint family, member 6 |
52760 |
0.13 |
| chr5_71506479_71506741 | 0.23 |
MAP1B |
microtubule-associated protein 1B |
31155 |
0.18 |
| chr2_23648910_23649061 | 0.23 |
KLHL29 |
kelch-like family member 29 |
40897 |
0.21 |
| chr10_121427681_121427947 | 0.23 |
BAG3 |
BCL2-associated athanogene 3 |
11692 |
0.22 |
| chr1_22196957_22197108 | 0.22 |
HSPG2 |
heparan sulfate proteoglycan 2 |
4167 |
0.23 |
| chr15_69850121_69850272 | 0.22 |
ENSG00000207119 |
. |
99708 |
0.06 |
| chr3_149825197_149825441 | 0.22 |
RP11-167H9.4 |
|
9500 |
0.22 |
| chr19_47730648_47731306 | 0.22 |
ENSG00000265134 |
. |
778 |
0.55 |
| chr1_227070649_227071204 | 0.22 |
PSEN2 |
presenilin 2 (Alzheimer disease 4) |
1398 |
0.48 |
| chr9_117048019_117048536 | 0.22 |
ORM1 |
orosomucoid 1 |
37059 |
0.14 |
| chr18_7608538_7608689 | 0.22 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
40796 |
0.2 |
| chr5_149537966_149538117 | 0.22 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
2618 |
0.22 |
| chr7_134511099_134511250 | 0.22 |
CALD1 |
caldesmon 1 |
17461 |
0.27 |
| chr1_156675699_156676565 | 0.22 |
CRABP2 |
cellular retinoic acid binding protein 2 |
524 |
0.6 |
| chr3_196453873_196454042 | 0.22 |
PAK2 |
p21 protein (Cdc42/Rac)-activated kinase 2 |
12771 |
0.11 |
| chr9_132465577_132465728 | 0.22 |
RP11-483H20.6 |
|
15262 |
0.14 |
| chr16_73248067_73248218 | 0.22 |
C16orf47 |
chromosome 16 open reading frame 47 |
69796 |
0.13 |
| chr9_97664466_97664617 | 0.22 |
RP11-49O14.2 |
|
1830 |
0.36 |
| chr12_63155759_63156101 | 0.21 |
ENSG00000200296 |
. |
88751 |
0.08 |
| chr9_74299304_74300206 | 0.21 |
TMEM2 |
transmembrane protein 2 |
20093 |
0.28 |
| chr8_134048854_134049005 | 0.21 |
TG |
thyroglobulin |
18985 |
0.21 |
| chrX_62878059_62878210 | 0.21 |
ARHGEF9-IT1 |
ARHGEF9 intronic transcript 1 (non-protein coding) |
13248 |
0.23 |
| chr9_14251173_14251324 | 0.21 |
NFIB |
nuclear factor I/B |
56764 |
0.15 |
| chr7_40509443_40509735 | 0.21 |
AC004988.1 |
|
76938 |
0.12 |
| chr1_61606862_61607139 | 0.21 |
RP4-802A10.1 |
|
16595 |
0.23 |
| chr10_21604717_21604868 | 0.21 |
ENSG00000207264 |
. |
6098 |
0.28 |
| chr22_18958063_18958554 | 0.21 |
DGCR5 |
DiGeorge syndrome critical region gene 5 (non-protein coding) |
212 |
0.93 |
| chr1_12466097_12466396 | 0.21 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
3678 |
0.3 |
| chr13_50976140_50976291 | 0.21 |
ENSG00000221198 |
. |
38842 |
0.22 |
| chr2_151375574_151375725 | 0.21 |
RND3 |
Rho family GTPase 3 |
19876 |
0.3 |
| chr4_169539578_169539729 | 0.21 |
PALLD |
palladin, cytoskeletal associated protein |
13113 |
0.21 |
| chr9_12818278_12818429 | 0.21 |
RP11-3L8.3 |
|
4009 |
0.26 |
| chr1_56540050_56540201 | 0.21 |
PIGQP1 |
phosphatidylinositol glycan anchor biosynthesis, class Q pseudogene 1 |
135172 |
0.05 |
| chr10_12746020_12746171 | 0.21 |
ENSG00000221331 |
. |
21257 |
0.25 |
| chr5_81683174_81683325 | 0.21 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
82083 |
0.11 |
| chr20_21164341_21164492 | 0.21 |
RP5-872K7.7 |
|
20885 |
0.19 |
| chr4_13919303_13919486 | 0.21 |
ENSG00000252092 |
. |
259143 |
0.02 |
| chr4_138580444_138580595 | 0.21 |
PCDH18 |
protocadherin 18 |
126871 |
0.06 |
| chr1_170278456_170278607 | 0.21 |
ENSG00000263384 |
. |
60905 |
0.14 |
| chr11_12854283_12854434 | 0.20 |
RP11-47J17.3 |
|
9144 |
0.19 |
| chr4_95411408_95411559 | 0.20 |
PDLIM5 |
PDZ and LIM domain 5 |
33393 |
0.24 |
| chr14_80064218_80064369 | 0.20 |
RP11-242P2.2 |
|
35361 |
0.24 |
| chr15_81482529_81482680 | 0.20 |
IL16 |
interleukin 16 |
6615 |
0.26 |
| chr1_201552534_201552685 | 0.20 |
AC096677.1 |
Uncharacterized protein ENSP00000471857 |
39404 |
0.12 |
| chr13_51891213_51891364 | 0.20 |
SERPINE3 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
18621 |
0.15 |
| chr1_236214774_236214925 | 0.20 |
ENSG00000252822 |
. |
9128 |
0.21 |
| chr1_162601274_162601425 | 0.20 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
186 |
0.95 |
| chr10_16552330_16552481 | 0.20 |
C1QL3 |
complement component 1, q subcomponent-like 3 |
11599 |
0.21 |
| chr1_25241073_25241224 | 0.20 |
RUNX3 |
runt-related transcription factor 3 |
14464 |
0.22 |
| chr20_60833882_60834033 | 0.20 |
OSBPL2 |
oxysterol binding protein-like 2 |
14408 |
0.14 |
| chr10_128352837_128352988 | 0.20 |
C10orf90 |
chromosome 10 open reading frame 90 |
6103 |
0.3 |
| chr6_129912108_129912259 | 0.20 |
RP1-69D17.3 |
|
109628 |
0.07 |
| chr6_128390231_128390382 | 0.20 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
4398 |
0.3 |
| chr9_82691298_82691449 | 0.20 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
369692 |
0.01 |
| chr5_83314054_83314205 | 0.20 |
ENSG00000216125 |
. |
44424 |
0.2 |
| chr7_134387393_134387662 | 0.20 |
CALD1 |
caldesmon 1 |
41476 |
0.17 |
| chr10_95984129_95984280 | 0.20 |
RP11-391J2.3 |
|
2841 |
0.29 |
| chr6_42223355_42223506 | 0.20 |
MRPS10 |
mitochondrial ribosomal protein S10 |
37827 |
0.15 |
| chr13_101045927_101046215 | 0.20 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
25318 |
0.23 |
| chr17_45955657_45955864 | 0.20 |
SP2 |
Sp2 transcription factor |
17756 |
0.1 |
| chr9_131337476_131337664 | 0.20 |
SPTAN1 |
spectrin, alpha, non-erythrocytic 1 |
22701 |
0.11 |
| chr15_101299874_101300025 | 0.20 |
ALDH1A3 |
aldehyde dehydrogenase 1 family, member A3 |
117970 |
0.05 |
| chr19_1361317_1362052 | 0.20 |
MUM1 |
melanoma associated antigen (mutated) 1 |
5321 |
0.1 |
| chr3_120163660_120163960 | 0.20 |
FSTL1 |
follistatin-like 1 |
6028 |
0.28 |
| chr9_117531894_117532045 | 0.20 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
22419 |
0.19 |
| chr21_43147797_43147948 | 0.20 |
LINC00112 |
long intergenic non-protein coding RNA 112 |
11276 |
0.18 |
| chr20_50073631_50073822 | 0.20 |
ENSG00000266761 |
. |
4212 |
0.31 |
| chr9_21685322_21685473 | 0.20 |
ENSG00000244230 |
. |
13916 |
0.23 |
| chr14_74257389_74257540 | 0.20 |
RP5-1021I20.1 |
|
1796 |
0.22 |
| chr1_56162477_56162628 | 0.20 |
ENSG00000272051 |
. |
211907 |
0.02 |
| chr5_97796152_97796303 | 0.20 |
ENSG00000223053 |
. |
178708 |
0.03 |
| chr8_119013250_119013401 | 0.20 |
EXT1 |
exostosin glycosyltransferase 1 |
109328 |
0.08 |
| chr2_160059536_160059687 | 0.20 |
WDSUB1 |
WD repeat, sterile alpha motif and U-box domain containing 1 |
83452 |
0.1 |
| chr11_36030745_36030896 | 0.20 |
ENSG00000263389 |
. |
828 |
0.67 |
| chr7_47824970_47825121 | 0.20 |
C7orf69 |
chromosome 7 open reading frame 69 |
9844 |
0.2 |
| chr14_52518580_52518731 | 0.20 |
NID2 |
nidogen 2 (osteonidogen) |
12985 |
0.2 |
| chr20_19796976_19797127 | 0.20 |
AL121761.2 |
Uncharacterized protein |
58372 |
0.12 |
| chr16_65580539_65580690 | 0.19 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
424346 |
0.01 |
| chr3_64211649_64211800 | 0.19 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
593 |
0.79 |
| chr1_161931528_161931772 | 0.19 |
OLFML2B |
olfactomedin-like 2B |
23372 |
0.22 |
| chr10_5637212_5637363 | 0.19 |
ENSG00000240577 |
. |
36139 |
0.13 |
| chr22_27856791_27856942 | 0.19 |
RP11-375H17.1 |
|
255602 |
0.02 |
| chr21_39668488_39669168 | 0.19 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
20 |
0.99 |
| chr3_50306894_50307086 | 0.19 |
SEMA3B-AS1 |
SEMA3B antisense RNA 1 (head to head) |
2187 |
0.14 |
| chr3_16225459_16225610 | 0.19 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
9318 |
0.25 |
| chr7_137222625_137222776 | 0.19 |
PTN |
pleiotrophin |
194089 |
0.03 |
| chr10_54247807_54248093 | 0.19 |
RP11-556E13.1 |
|
72915 |
0.13 |
| chr5_133330291_133330536 | 0.19 |
VDAC1 |
voltage-dependent anion channel 1 |
9913 |
0.22 |
| chr11_111782921_111783072 | 0.19 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
30 |
0.76 |
| chr17_13497728_13497879 | 0.19 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
7441 |
0.27 |
| chr1_94730956_94731107 | 0.19 |
ARHGAP29 |
Rho GTPase activating protein 29 |
27842 |
0.21 |
| chr14_77451867_77452087 | 0.19 |
ENSG00000266553 |
. |
17869 |
0.19 |
| chr13_51104174_51104472 | 0.19 |
ENSG00000221198 |
. |
166950 |
0.04 |
| chr9_33131864_33132111 | 0.19 |
B4GALT1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
3790 |
0.2 |
| chr8_123445640_123445791 | 0.19 |
ENSG00000238901 |
. |
237815 |
0.02 |
| chr2_44461275_44461426 | 0.19 |
ENSG00000239052 |
. |
5450 |
0.2 |
| chr9_35277809_35277960 | 0.19 |
AL160274.1 |
HCG17281; PRO0038; Uncharacterized protein |
83378 |
0.07 |
| chr12_68189071_68189222 | 0.19 |
RP11-335O4.3 |
|
124447 |
0.06 |
| chr3_99975378_99975668 | 0.19 |
TBC1D23 |
TBC1 domain family, member 23 |
4321 |
0.22 |
| chr11_86661756_86661907 | 0.19 |
FZD4 |
frizzled family receptor 4 |
4602 |
0.27 |
| chr12_53442686_53442837 | 0.19 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
28 |
0.95 |
| chr5_14039644_14039795 | 0.19 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
95067 |
0.09 |
| chr13_76196300_76196451 | 0.19 |
LMO7 |
LIM domain 7 |
1805 |
0.32 |
| chr14_102449329_102449480 | 0.19 |
DYNC1H1 |
dynein, cytoplasmic 1, heavy chain 1 |
18539 |
0.19 |
| chr2_203000046_203000197 | 0.19 |
AC079354.3 |
|
18340 |
0.14 |
| chr1_31480914_31481065 | 0.19 |
PUM1 |
pumilio RNA-binding family member 1 |
13038 |
0.19 |
| chr20_51693240_51693391 | 0.19 |
ENSG00000252629 |
. |
14159 |
0.21 |
| chr6_56616320_56616471 | 0.19 |
DST |
dystonin |
34282 |
0.21 |
| chr8_89336416_89336710 | 0.19 |
RP11-586K2.1 |
|
2502 |
0.31 |
| chr19_10529958_10530965 | 0.19 |
CDC37 |
cell division cycle 37 |
336 |
0.7 |
| chr3_197680241_197680392 | 0.19 |
RPL35A |
ribosomal protein L35a |
2888 |
0.19 |
| chr12_47996581_47996732 | 0.18 |
RPAP3 |
RNA polymerase II associated protein 3 |
103113 |
0.06 |
| chr20_50060887_50061038 | 0.18 |
ENSG00000266761 |
. |
8552 |
0.27 |
| chr5_67615920_67616071 | 0.18 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
27599 |
0.25 |
| chr5_82863496_82863647 | 0.18 |
VCAN-AS1 |
VCAN antisense RNA 1 |
5438 |
0.29 |
| chr1_60061425_60061576 | 0.18 |
FGGY |
FGGY carbohydrate kinase domain containing |
41910 |
0.2 |
| chr22_37945927_37946078 | 0.18 |
RP5-1177I5.3 |
|
31 |
0.96 |
| chr9_114821454_114821605 | 0.18 |
RP11-4O1.2 |
|
21519 |
0.18 |
| chr6_132272833_132273094 | 0.18 |
CTGF |
connective tissue growth factor |
450 |
0.69 |
| chr2_67790952_67791103 | 0.18 |
ETAA1 |
Ewing tumor-associated antigen 1 |
166576 |
0.04 |
| chr20_38429169_38429320 | 0.18 |
ENSG00000263518 |
. |
302754 |
0.01 |
| chr1_201505428_201505649 | 0.18 |
CSRP1 |
cysteine and glycine-rich protein 1 |
26954 |
0.14 |
| chr7_18809523_18809674 | 0.18 |
ENSG00000222164 |
. |
38304 |
0.21 |
| chr8_42389920_42390219 | 0.18 |
RP11-503E24.2 |
|
3378 |
0.21 |
| chr3_139028342_139028493 | 0.18 |
MRPS22 |
mitochondrial ribosomal protein S22 |
34444 |
0.14 |
| chr7_100200945_100201346 | 0.18 |
PCOLCE-AS1 |
PCOLCE antisense RNA 1 |
487 |
0.56 |
| chr15_30205727_30205878 | 0.18 |
TJP1 |
tight junction protein 1 |
55266 |
0.13 |
| chr20_46081515_46081666 | 0.18 |
ENSG00000202186 |
. |
31535 |
0.14 |
| chr8_327162_327457 | 0.18 |
FBXO25 |
F-box protein 25 |
29119 |
0.2 |
| chr3_131853729_131854016 | 0.18 |
CPNE4 |
copine IV |
95422 |
0.08 |
| chr1_185943240_185943391 | 0.18 |
HMCN1 |
hemicentin 1 |
197204 |
0.03 |
| chr6_114316634_114316785 | 0.18 |
RP3-399L15.3 |
|
301 |
0.91 |
| chr5_158487783_158487934 | 0.18 |
EBF1 |
early B-cell factor 1 |
38843 |
0.17 |
| chr6_52589214_52589365 | 0.18 |
GSTA2 |
glutathione S-transferase alpha 2 |
39078 |
0.14 |
| chr17_48173021_48173384 | 0.18 |
PDK2 |
pyruvate dehydrogenase kinase, isozyme 2 |
136 |
0.93 |
| chr1_68797213_68797364 | 0.18 |
WLS |
wntless Wnt ligand secretion mediator |
98485 |
0.07 |
| chr13_80511305_80511456 | 0.18 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
402414 |
0.01 |
| chr6_100624567_100624718 | 0.18 |
MCHR2-AS1 |
MCHR2 antisense RNA 1 |
175826 |
0.03 |
| chr1_16479115_16479478 | 0.18 |
RP11-276H7.2 |
|
2410 |
0.18 |
| chr5_111071307_111071458 | 0.18 |
STARD4-AS1 |
STARD4 antisense RNA 1 |
5296 |
0.22 |
| chr3_64307670_64307821 | 0.18 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
54090 |
0.15 |
| chr2_85390403_85390554 | 0.18 |
TCF7L1-IT1 |
TCF7L1 intronic transcript 1 (non-protein coding) |
23054 |
0.15 |
| chr12_113669340_113669491 | 0.18 |
TPCN1 |
two pore segment channel 1 |
6366 |
0.14 |
| chr1_23891322_23891473 | 0.18 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
5112 |
0.2 |
| chr2_9239281_9239456 | 0.18 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
95426 |
0.08 |
| chr7_865029_865180 | 0.18 |
SUN1 |
Sad1 and UNC84 domain containing 1 |
5463 |
0.2 |
| chr3_115042385_115042536 | 0.18 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
176342 |
0.03 |
| chr12_107372094_107372245 | 0.18 |
MTERFD3 |
MTERF domain containing 3 |
6895 |
0.17 |
| chr10_88729130_88729281 | 0.18 |
MMRN2 |
multimerin 2 |
33 |
0.75 |
| chr10_28584404_28584657 | 0.18 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
7465 |
0.24 |
| chr2_101383857_101384008 | 0.18 |
NPAS2 |
neuronal PAS domain protein 2 |
52682 |
0.14 |
| chr9_132431325_132431476 | 0.18 |
PRRX2 |
paired related homeobox 2 |
3480 |
0.17 |
| chr6_100624308_100624459 | 0.18 |
MCHR2-AS1 |
MCHR2 antisense RNA 1 |
175567 |
0.03 |
| chr4_7873753_7873904 | 0.18 |
AFAP1 |
actin filament associated protein 1 |
19 |
0.98 |
| chr11_27238222_27238373 | 0.18 |
RP11-1L12.3 |
|
1985 |
0.47 |
| chr2_145235114_145235402 | 0.18 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
39857 |
0.18 |
| chr2_187459352_187459503 | 0.18 |
ITGAV |
integrin, alpha V |
4620 |
0.27 |
| chr5_133883689_133883848 | 0.18 |
JADE2 |
jade family PHD finger 2 |
21441 |
0.15 |
| chr17_75127867_75128287 | 0.18 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
4122 |
0.23 |
| chr1_113070330_113070481 | 0.18 |
RP4-671G15.2 |
|
9342 |
0.18 |
| chr3_59797932_59798083 | 0.17 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
159576 |
0.04 |
| chr13_43681816_43681967 | 0.17 |
DNAJC15 |
DnaJ (Hsp40) homolog, subfamily C, member 15 |
84552 |
0.1 |
| chr17_2085515_2085666 | 0.17 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
9118 |
0.12 |
| chr3_183947384_183948074 | 0.17 |
VWA5B2 |
von Willebrand factor A domain containing 5B2 |
488 |
0.65 |
| chr11_12710648_12710799 | 0.17 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
13673 |
0.28 |
| chr1_115857199_115857350 | 0.17 |
NGF |
nerve growth factor (beta polypeptide) |
23583 |
0.21 |
| chr6_38306218_38306369 | 0.17 |
ENSG00000238716 |
. |
20712 |
0.26 |
| chr17_20932042_20932193 | 0.17 |
USP22 |
ubiquitin specific peptidase 22 |
9665 |
0.18 |
| chr1_35972126_35972305 | 0.17 |
KIAA0319L |
KIAA0319-like |
253 |
0.92 |
| chr7_24865334_24866225 | 0.17 |
DFNA5 |
deafness, autosomal dominant 5 |
56535 |
0.13 |
| chr4_5723966_5724117 | 0.17 |
EVC |
Ellis van Creveld syndrome |
11099 |
0.23 |
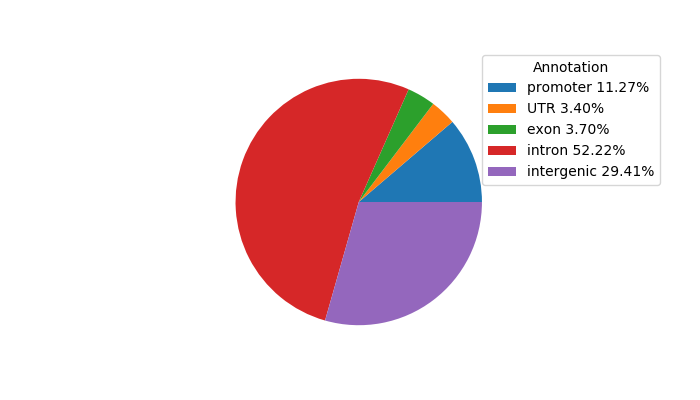
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 0.2 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.2 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.2 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.3 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis(GO:0060502) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0090559 | regulation of mitochondrial membrane permeability(GO:0046902) regulation of membrane permeability(GO:0090559) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0030540 | female genitalia development(GO:0030540) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) negative regulation of lipase activity(GO:0060192) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0010510 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.0 | GO:0051532 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.0 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.0 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0006577 | amino-acid betaine metabolic process(GO:0006577) carnitine metabolic process(GO:0009437) |
| 0.0 | 0.0 | GO:0070828 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.0 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |