Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
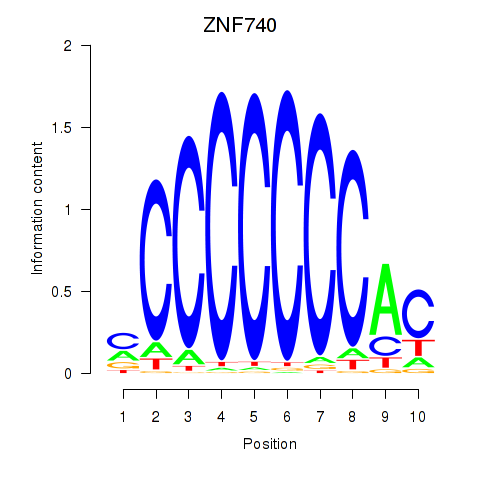
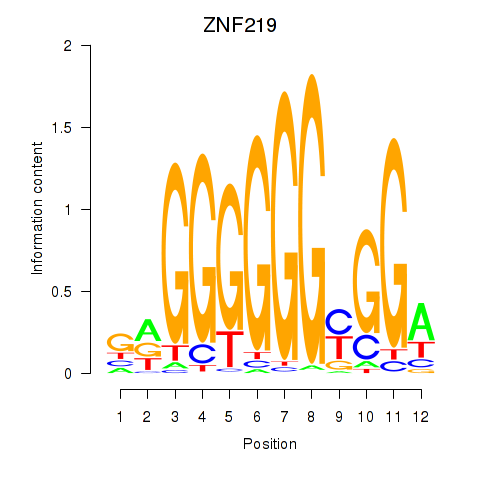
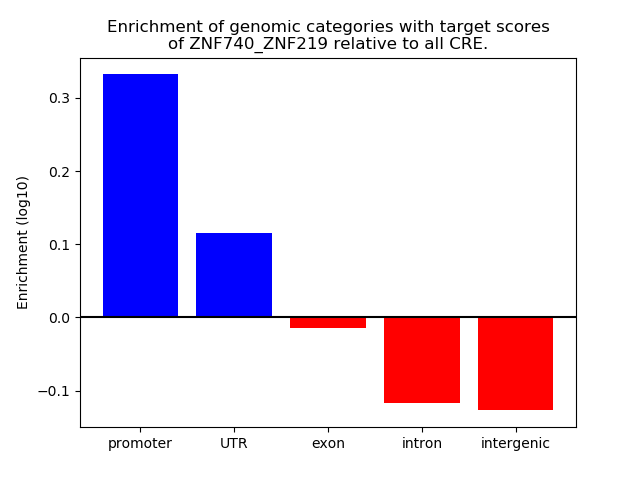
Results for ZNF740_ZNF219
Z-value: 0.97
Transcription factors associated with ZNF740_ZNF219
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF740
|
ENSG00000139651.9 | zinc finger protein 740 |
|
ZNF219
|
ENSG00000165804.11 | zinc finger protein 219 |
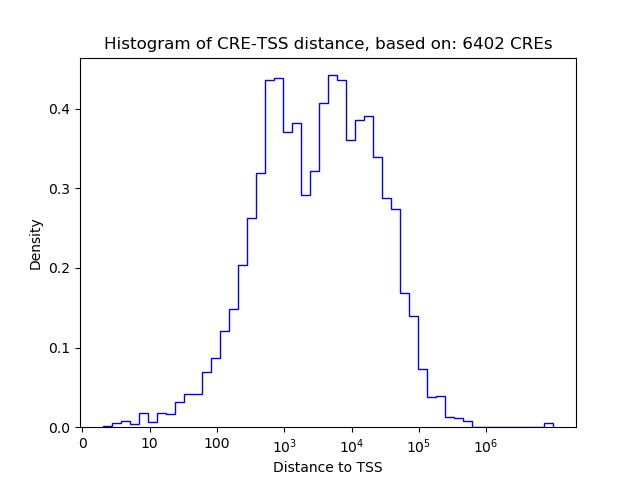
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_21567463_21567799 | ZNF219 | 458 | 0.454825 | -0.71 | 3.1e-02 | Click! |
| chr14_21560976_21561143 | ZNF219 | 928 | 0.371417 | 0.68 | 4.3e-02 | Click! |
| chr14_21557704_21557855 | ZNF219 | 4208 | 0.106934 | 0.66 | 5.4e-02 | Click! |
| chr14_21556036_21556544 | ZNF219 | 5697 | 0.096919 | 0.52 | 1.5e-01 | Click! |
| chr14_21552997_21553148 | ZNF219 | 8915 | 0.087630 | 0.43 | 2.4e-01 | Click! |
Activity of the ZNF740_ZNF219 motif across conditions
Conditions sorted by the z-value of the ZNF740_ZNF219 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
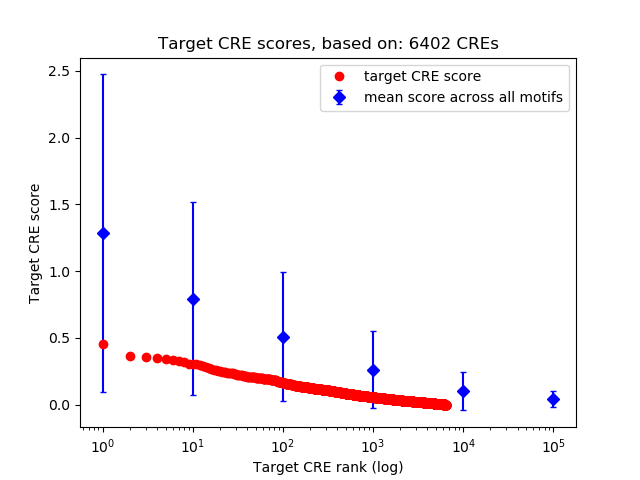
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_11203608_11204588 | 0.46 |
CELF2 |
CUGBP, Elav-like family member 2 |
2895 |
0.29 |
| chr5_175916247_175916556 | 0.36 |
ENSG00000221373 |
. |
37638 |
0.09 |
| chr5_75680037_75680677 | 0.36 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
18717 |
0.25 |
| chr15_31657045_31657355 | 0.35 |
KLF13 |
Kruppel-like factor 13 |
1157 |
0.65 |
| chrX_102940963_102941114 | 0.34 |
MORF4L2 |
mortality factor 4 like 2 |
111 |
0.93 |
| chr1_27929391_27929542 | 0.33 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
636 |
0.66 |
| chr10_126849591_126849742 | 0.33 |
CTBP2 |
C-terminal binding protein 2 |
36 |
0.99 |
| chr2_134884979_134885130 | 0.32 |
ENSG00000263813 |
. |
358 |
0.9 |
| chr17_76122728_76122959 | 0.31 |
TMC6 |
transmembrane channel-like 6 |
258 |
0.86 |
| chr11_66046306_66046457 | 0.31 |
CNIH2 |
cornichon family AMPA receptor auxiliary protein 2 |
711 |
0.33 |
| chr2_99377617_99378126 | 0.30 |
ENSG00000201070 |
. |
21002 |
0.19 |
| chr18_22931267_22931732 | 0.30 |
ZNF521 |
zinc finger protein 521 |
80 |
0.98 |
| chr2_106459235_106459386 | 0.29 |
NCK2 |
NCK adaptor protein 2 |
8894 |
0.24 |
| chr15_77305575_77305811 | 0.28 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
2459 |
0.29 |
| chr11_68295708_68295914 | 0.27 |
PPP6R3 |
protein phosphatase 6, regulatory subunit 3 |
7068 |
0.19 |
| chr18_77280687_77280838 | 0.27 |
AC018445.1 |
Uncharacterized protein |
4705 |
0.31 |
| chr21_44204010_44204161 | 0.26 |
AP001627.1 |
|
42217 |
0.15 |
| chr3_13026960_13027112 | 0.26 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
1500 |
0.52 |
| chr8_144717528_144717700 | 0.25 |
ZNF623 |
zinc finger protein 623 |
569 |
0.48 |
| chr10_14641210_14641531 | 0.25 |
FAM107B |
family with sequence similarity 107, member B |
5018 |
0.26 |
| chr2_136885184_136885335 | 0.25 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
9524 |
0.29 |
| chr3_178787798_178787973 | 0.24 |
ZMAT3 |
zinc finger, matrin-type 3 |
1469 |
0.45 |
| chr17_72743484_72743750 | 0.24 |
ENSG00000264624 |
. |
1135 |
0.23 |
| chr3_17774733_17775005 | 0.24 |
TBC1D5 |
TBC1 domain family, member 5 |
7530 |
0.31 |
| chr7_19155701_19156072 | 0.24 |
TWIST1 |
twist family bHLH transcription factor 1 |
1409 |
0.34 |
| chr14_99730566_99730874 | 0.24 |
AL109767.1 |
|
1435 |
0.45 |
| chr19_13025260_13025411 | 0.24 |
SYCE2 |
synaptonemal complex central element protein 2 |
4755 |
0.07 |
| chr12_72055858_72056376 | 0.24 |
THAP2 |
THAP domain containing, apoptosis associated protein 2 |
672 |
0.58 |
| chr2_113402705_113402940 | 0.23 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
612 |
0.75 |
| chr18_77282001_77282189 | 0.23 |
AC018445.1 |
Uncharacterized protein |
6038 |
0.3 |
| chr9_92092219_92092635 | 0.23 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
2378 |
0.35 |
| chr1_156629969_156630120 | 0.22 |
RP11-284F21.7 |
|
1172 |
0.28 |
| chr19_2303224_2303375 | 0.22 |
LINGO3 |
leucine rich repeat and Ig domain containing 3 |
4857 |
0.1 |
| chr9_97022637_97022801 | 0.22 |
ZNF169 |
zinc finger protein 169 |
1109 |
0.46 |
| chr8_27198953_27199192 | 0.22 |
PTK2B |
protein tyrosine kinase 2 beta |
14732 |
0.2 |
| chr18_77280955_77281122 | 0.22 |
AC018445.1 |
Uncharacterized protein |
4981 |
0.31 |
| chr1_79111221_79111724 | 0.22 |
IFI44 |
interferon-induced protein 44 |
4044 |
0.25 |
| chr1_154985178_154985329 | 0.21 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
1671 |
0.16 |
| chr13_31313160_31313326 | 0.21 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
3598 |
0.34 |
| chr1_167415328_167415582 | 0.21 |
RP11-104L21.2 |
|
12443 |
0.21 |
| chr16_50700983_50701245 | 0.21 |
RP11-401P9.5 |
|
992 |
0.45 |
| chr16_81815354_81815505 | 0.21 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
2566 |
0.4 |
| chr1_154465561_154465817 | 0.21 |
SHE |
Src homology 2 domain containing E |
4047 |
0.14 |
| chr10_61659889_61660066 | 0.21 |
CCDC6 |
coiled-coil domain containing 6 |
6437 |
0.28 |
| chr2_8444024_8444411 | 0.21 |
AC011747.7 |
|
371679 |
0.01 |
| chr19_33769711_33769869 | 0.21 |
CTD-2540B15.11 |
|
21050 |
0.11 |
| chr1_84326924_84327075 | 0.21 |
ENSG00000223231 |
. |
67439 |
0.13 |
| chr14_21082227_21082378 | 0.20 |
RNASE11 |
ribonuclease, RNase A family, 11 (non-active) |
4259 |
0.13 |
| chr3_52930379_52930530 | 0.20 |
TMEM110-MUSTN1 |
TMEM110-MUSTN1 readthrough |
1124 |
0.33 |
| chr22_19752877_19753143 | 0.20 |
TBX1 |
T-box 1 |
8784 |
0.18 |
| chr5_137429102_137429305 | 0.20 |
WNT8A |
wingless-type MMTV integration site family, member 8A |
9429 |
0.11 |
| chr7_77564423_77564574 | 0.20 |
ENSG00000222432 |
. |
39301 |
0.17 |
| chr4_146542708_146542859 | 0.20 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
3368 |
0.26 |
| chr1_200987046_200987749 | 0.20 |
KIF21B |
kinesin family member 21B |
5139 |
0.21 |
| chr10_131769782_131769953 | 0.20 |
EBF3 |
early B-cell factor 3 |
7762 |
0.31 |
| chr17_29820821_29821086 | 0.20 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
5827 |
0.15 |
| chr22_36801197_36801348 | 0.20 |
MYH9 |
myosin, heavy chain 9, non-muscle |
17209 |
0.15 |
| chr19_7744064_7744215 | 0.20 |
CTD-3214H19.16 |
|
752 |
0.37 |
| chr2_109241859_109242070 | 0.20 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
4242 |
0.27 |
| chr4_78722788_78722939 | 0.20 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
17354 |
0.25 |
| chr9_96229432_96229584 | 0.20 |
FAM120AOS |
family with sequence similarity 120A opposite strand |
13634 |
0.14 |
| chr10_14919889_14920040 | 0.20 |
SUV39H2 |
suppressor of variegation 3-9 homolog 2 (Drosophila) |
883 |
0.58 |
| chr9_102855874_102856025 | 0.19 |
ERP44 |
endoplasmic reticulum protein 44 |
5373 |
0.2 |
| chr9_126030120_126030336 | 0.19 |
STRBP |
spermatid perinuclear RNA binding protein |
615 |
0.81 |
| chr1_151802545_151802942 | 0.19 |
RORC |
RAR-related orphan receptor C |
1483 |
0.2 |
| chr22_20784316_20784467 | 0.19 |
SCARF2 |
scavenger receptor class F, member 2 |
7721 |
0.11 |
| chr3_191047673_191047824 | 0.19 |
UTS2B |
urotensin 2B |
514 |
0.64 |
| chrX_100664537_100664740 | 0.19 |
HNRNPH2 |
heterogeneous nuclear ribonucleoprotein H2 (H') |
1355 |
0.25 |
| chr17_74269143_74269364 | 0.19 |
QRICH2 |
glutamine rich 2 |
7185 |
0.13 |
| chrX_44768647_44768798 | 0.19 |
ENSG00000252113 |
. |
23971 |
0.2 |
| chr10_8094760_8094911 | 0.19 |
GATA3 |
GATA binding protein 3 |
1821 |
0.52 |
| chr1_234349337_234349623 | 0.19 |
SLC35F3 |
solute carrier family 35, member F3 |
537 |
0.68 |
| chr9_139482691_139482911 | 0.19 |
ENSG00000252440 |
. |
14166 |
0.1 |
| chr8_27181557_27182090 | 0.19 |
PTK2B |
protein tyrosine kinase 2 beta |
1039 |
0.56 |
| chr1_155036684_155036860 | 0.19 |
EFNA4 |
ephrin-A4 |
535 |
0.37 |
| chr22_30582834_30583084 | 0.19 |
RP3-438O4.4 |
|
20139 |
0.13 |
| chrX_128908006_128908241 | 0.19 |
SASH3 |
SAM and SH3 domain containing 3 |
5837 |
0.21 |
| chr14_91859539_91859984 | 0.18 |
CCDC88C |
coiled-coil domain containing 88C |
23929 |
0.2 |
| chr17_7789286_7789521 | 0.18 |
LSMD1 |
LSM domain containing 1 |
876 |
0.33 |
| chr8_38324368_38324696 | 0.18 |
FGFR1 |
fibroblast growth factor receptor 1 |
319 |
0.88 |
| chr11_2326434_2326626 | 0.18 |
TSPAN32 |
tetraspanin 32 |
2432 |
0.18 |
| chr14_91873415_91873761 | 0.18 |
CCDC88C |
coiled-coil domain containing 88C |
10102 |
0.24 |
| chr2_27615435_27615612 | 0.18 |
ZNF513 |
zinc finger protein 513 |
11866 |
0.08 |
| chr17_79420586_79420890 | 0.18 |
ENSG00000266189 |
. |
2524 |
0.16 |
| chr17_29819355_29819635 | 0.18 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
4369 |
0.16 |
| chr1_167408168_167408319 | 0.18 |
RP11-104L21.2 |
|
19655 |
0.19 |
| chr8_128746908_128747308 | 0.17 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
572 |
0.84 |
| chr12_133055280_133055431 | 0.17 |
MUC8 |
mucin 8 |
4629 |
0.23 |
| chr6_90868295_90868522 | 0.17 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
138053 |
0.04 |
| chr11_64012927_64013269 | 0.17 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
190 |
0.61 |
| chr2_145275512_145275663 | 0.17 |
ZEB2-AS1 |
ZEB2 antisense RNA 1 |
77 |
0.56 |
| chr14_100525569_100526087 | 0.17 |
EVL |
Enah/Vasp-like |
5787 |
0.17 |
| chr1_26612161_26612476 | 0.17 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
5705 |
0.12 |
| chr17_66251665_66252057 | 0.17 |
ARSG |
arylsulfatase G |
3462 |
0.19 |
| chr15_78330161_78330463 | 0.17 |
ENSG00000221476 |
. |
561 |
0.69 |
| chr17_66203318_66203469 | 0.17 |
AMZ2 |
archaelysin family metallopeptidase 2 |
40322 |
0.12 |
| chr17_39941615_39941766 | 0.17 |
JUP |
junction plakoglobin |
283 |
0.83 |
| chr1_198490580_198490731 | 0.17 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
19149 |
0.27 |
| chr7_2739159_2739434 | 0.16 |
AMZ1 |
archaelysin family metallopeptidase 1 |
11460 |
0.21 |
| chr8_22299308_22299495 | 0.16 |
PPP3CC |
protein phosphatase 3, catalytic subunit, gamma isozyme |
523 |
0.74 |
| chr15_69075380_69075658 | 0.16 |
ENSG00000265195 |
. |
18745 |
0.22 |
| chr14_64966305_64966456 | 0.16 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
4063 |
0.14 |
| chr16_49892506_49892687 | 0.16 |
ZNF423 |
zinc finger protein 423 |
766 |
0.73 |
| chr22_36557619_36557770 | 0.16 |
APOL3 |
apolipoprotein L, 3 |
717 |
0.72 |
| chr4_154417485_154417761 | 0.16 |
KIAA0922 |
KIAA0922 |
30122 |
0.21 |
| chr7_949490_949865 | 0.16 |
ADAP1 |
ArfGAP with dual PH domains 1 |
4963 |
0.16 |
| chr19_16475924_16476075 | 0.16 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
3235 |
0.2 |
| chr11_128600956_128601302 | 0.16 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
33556 |
0.15 |
| chr3_183980771_183980922 | 0.16 |
CAMK2N2 |
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
1595 |
0.22 |
| chr6_17601986_17602159 | 0.16 |
FAM8A1 |
family with sequence similarity 8, member A1 |
1486 |
0.44 |
| chr1_243622637_243622945 | 0.16 |
RP11-269F20.1 |
|
86043 |
0.09 |
| chr3_71287500_71287651 | 0.16 |
FOXP1 |
forkhead box P1 |
6741 |
0.29 |
| chr1_902652_902803 | 0.15 |
PLEKHN1 |
pleckstrin homology domain containing, family N member 1 |
845 |
0.39 |
| chr18_57025739_57025916 | 0.15 |
LMAN1 |
lectin, mannose-binding, 1 |
1367 |
0.48 |
| chr17_15495904_15496128 | 0.15 |
CDRT1 |
CMT1A duplicated region transcript 1 |
706 |
0.58 |
| chr13_99993452_99993845 | 0.15 |
ENSG00000207719 |
. |
14737 |
0.19 |
| chr5_176514554_176514864 | 0.15 |
FGFR4 |
fibroblast growth factor receptor 4 |
289 |
0.9 |
| chr5_170815911_170816235 | 0.15 |
NPM1 |
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
1188 |
0.38 |
| chr17_39930444_39930595 | 0.15 |
JUP |
junction plakoglobin |
1998 |
0.19 |
| chr7_3152682_3152883 | 0.15 |
AC091801.1 |
LOC392621; Uncharacterized protein |
45106 |
0.16 |
| chr5_159932695_159932967 | 0.15 |
ENSG00000253522 |
. |
20472 |
0.17 |
| chr11_68530734_68530885 | 0.15 |
MTL5 |
metallothionein-like 5, testis-specific (tesmin) |
11777 |
0.2 |
| chr17_79005380_79005531 | 0.15 |
BAIAP2 |
BAI1-associated protein 2 |
3495 |
0.16 |
| chr16_30935176_30935327 | 0.15 |
FBXL19 |
F-box and leucine-rich repeat protein 19 |
167 |
0.8 |
| chr14_65878762_65878997 | 0.15 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
225 |
0.95 |
| chr19_45738680_45738986 | 0.15 |
EXOC3L2 |
exocyst complex component 3-like 2 |
1364 |
0.32 |
| chr22_39487886_39488384 | 0.15 |
RP4-742C19.12 |
|
544 |
0.68 |
| chr2_227663405_227663664 | 0.15 |
IRS1 |
insulin receptor substrate 1 |
941 |
0.59 |
| chr1_109816776_109816927 | 0.15 |
PSRC1 |
proline/serine-rich coiled-coil 1 |
7450 |
0.15 |
| chr3_46244229_46244380 | 0.15 |
CCR1 |
chemokine (C-C motif) receptor 1 |
5583 |
0.25 |
| chr13_41139475_41139713 | 0.15 |
AL133318.1 |
Uncharacterized protein |
28271 |
0.22 |
| chr5_133437895_133438046 | 0.15 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
12432 |
0.22 |
| chr3_38039585_38039999 | 0.15 |
VILL |
villin-like |
4148 |
0.16 |
| chr1_33282419_33282585 | 0.15 |
S100PBP |
S100P binding protein |
134 |
0.93 |
| chr17_40937018_40937222 | 0.14 |
WNK4 |
WNK lysine deficient protein kinase 4 |
4424 |
0.09 |
| chr5_180338131_180338282 | 0.14 |
ENSG00000222852 |
. |
1368 |
0.33 |
| chr19_1609011_1609503 | 0.14 |
UQCR11 |
ubiquinol-cytochrome c reductase, complex III subunit XI |
3777 |
0.11 |
| chr17_55681912_55682272 | 0.14 |
RP11-118E18.4 |
|
3681 |
0.24 |
| chr17_44229938_44230089 | 0.14 |
KANSL1 |
KAT8 regulatory NSL complex subunit 1 |
19530 |
0.15 |
| chr16_81554932_81555083 | 0.14 |
CMIP |
c-Maf inducing protein |
26053 |
0.24 |
| chr3_139107166_139107509 | 0.14 |
ENSG00000252245 |
. |
218 |
0.84 |
| chr2_239323617_239323768 | 0.14 |
ENSG00000202099 |
. |
2409 |
0.26 |
| chr18_13612636_13612881 | 0.14 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
140 |
0.93 |
| chrX_30589733_30589884 | 0.14 |
CXorf21 |
chromosome X open reading frame 21 |
6153 |
0.27 |
| chr17_79318286_79318703 | 0.14 |
TMEM105 |
transmembrane protein 105 |
14020 |
0.13 |
| chr16_53421654_53421805 | 0.14 |
RP11-44F14.1 |
|
14734 |
0.16 |
| chr11_119545202_119545353 | 0.14 |
ENSG00000199217 |
. |
18256 |
0.18 |
| chr1_205243235_205243736 | 0.14 |
TMCC2 |
transmembrane and coiled-coil domain family 2 |
18156 |
0.15 |
| chr2_64835263_64835414 | 0.14 |
ENSG00000252414 |
. |
29312 |
0.17 |
| chrX_78337599_78337998 | 0.14 |
GPR174 |
G protein-coupled receptor 174 |
88671 |
0.1 |
| chr8_11313733_11313920 | 0.14 |
FAM167A |
family with sequence similarity 167, member A |
10450 |
0.15 |
| chr14_99950613_99950764 | 0.14 |
CCNK |
cyclin K |
2926 |
0.2 |
| chr1_23856240_23856391 | 0.14 |
E2F2 |
E2F transcription factor 2 |
1397 |
0.41 |
| chr10_21805327_21805676 | 0.14 |
SKIDA1 |
SKI/DACH domain containing 1 |
1347 |
0.33 |
| chr19_17637669_17637829 | 0.14 |
FAM129C |
family with sequence similarity 129, member C |
298 |
0.81 |
| chr20_61558751_61558930 | 0.14 |
DIDO1 |
death inducer-obliterator 1 |
985 |
0.48 |
| chr2_85166364_85166744 | 0.14 |
KCMF1 |
potassium channel modulatory factor 1 |
31662 |
0.15 |
| chrX_30848915_30849066 | 0.14 |
TAB3-AS1 |
TAB3 antisense RNA 1 |
3750 |
0.23 |
| chr1_10753317_10753525 | 0.14 |
CASZ1 |
castor zinc finger 1 |
48288 |
0.13 |
| chr16_28156476_28156809 | 0.14 |
XPO6 |
exportin 6 |
24580 |
0.16 |
| chr10_102982257_102982408 | 0.14 |
LBX1-AS1 |
LBX1 antisense RNA 1 (head to head) |
7019 |
0.15 |
| chr14_93798178_93798526 | 0.14 |
BTBD7 |
BTB (POZ) domain containing 7 |
340 |
0.8 |
| chr14_24046924_24047247 | 0.14 |
JPH4 |
junctophilin 4 |
909 |
0.42 |
| chr1_247693623_247693886 | 0.14 |
OR2C3 |
olfactory receptor, family 2, subfamily C, member 3 |
3387 |
0.15 |
| chr1_236051628_236051779 | 0.14 |
LYST |
lysosomal trafficking regulator |
4831 |
0.19 |
| chr2_95719306_95719457 | 0.13 |
AC103563.9 |
|
460 |
0.79 |
| chr15_79383402_79383553 | 0.13 |
RASGRF1 |
Ras protein-specific guanine nucleotide-releasing factor 1 |
362 |
0.89 |
| chrX_48648064_48648437 | 0.13 |
GATA1 |
GATA binding protein 1 (globin transcription factor 1) |
3269 |
0.13 |
| chr6_36169985_36170141 | 0.13 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
1989 |
0.28 |
| chr9_129326429_129326580 | 0.13 |
ENSG00000221173 |
. |
12405 |
0.18 |
| chr7_151215555_151215799 | 0.13 |
RHEB |
Ras homolog enriched in brain |
522 |
0.78 |
| chr5_180632651_180632802 | 0.13 |
CTC-338M12.1 |
|
394 |
0.43 |
| chr19_54378263_54378435 | 0.13 |
AC008440.5 |
|
954 |
0.29 |
| chr2_242802197_242802348 | 0.13 |
PDCD1 |
programmed cell death 1 |
1212 |
0.32 |
| chr3_114865147_114865298 | 0.13 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
896 |
0.76 |
| chr9_95857394_95857545 | 0.13 |
RP11-274J16.5 |
|
342 |
0.77 |
| chr5_176924121_176924335 | 0.13 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
334 |
0.76 |
| chr17_79140382_79140553 | 0.13 |
AATK |
apoptosis-associated tyrosine kinase |
650 |
0.45 |
| chr1_24645534_24645796 | 0.13 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
147 |
0.95 |
| chr12_6714982_6715407 | 0.13 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
652 |
0.48 |
| chr3_55521660_55522023 | 0.13 |
WNT5A-AS1 |
WNT5A antisense RNA 1 |
114 |
0.87 |
| chr9_126108320_126108471 | 0.13 |
CRB2 |
crumbs homolog 2 (Drosophila) |
10054 |
0.24 |
| chr11_30346022_30346188 | 0.13 |
ARL14EP |
ADP-ribosylation factor-like 14 effector protein |
1507 |
0.54 |
| chr22_30683089_30683240 | 0.13 |
GATSL3 |
GATS protein-like 3 |
2432 |
0.15 |
| chr9_33817398_33817619 | 0.13 |
UBE2R2 |
ubiquitin-conjugating enzyme E2R 2 |
57 |
0.94 |
| chrX_39920814_39920965 | 0.13 |
BCOR |
BCL6 corepressor |
1301 |
0.62 |
| chr19_40973350_40973501 | 0.13 |
SPTBN4 |
spectrin, beta, non-erythrocytic 4 |
299 |
0.81 |
| chr5_55982532_55982749 | 0.13 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
80581 |
0.09 |
| chr4_2965787_2965938 | 0.13 |
GRK4 |
G protein-coupled receptor kinase 4 |
274 |
0.77 |
| chr2_143871194_143871345 | 0.13 |
ARHGAP15 |
Rho GTPase activating protein 15 |
15614 |
0.25 |
| chr2_96811667_96811891 | 0.13 |
DUSP2 |
dual specificity phosphatase 2 |
600 |
0.68 |
| chr8_81070516_81070667 | 0.13 |
TPD52 |
tumor protein D52 |
12844 |
0.2 |
| chr12_124762466_124762617 | 0.13 |
FAM101A |
family with sequence similarity 101, member A |
11169 |
0.25 |
| chr1_26697390_26697759 | 0.13 |
ZNF683 |
zinc finger protein 683 |
333 |
0.82 |
| chr4_8231324_8231505 | 0.13 |
SH3TC1 |
SH3 domain and tetratricopeptide repeats 1 |
11157 |
0.22 |
| chr2_172105282_172105509 | 0.13 |
TLK1 |
tousled-like kinase 1 |
17571 |
0.25 |
| chr20_50016749_50017039 | 0.13 |
ENSG00000263645 |
. |
23036 |
0.23 |
| chr1_77748623_77748894 | 0.13 |
AK5 |
adenylate kinase 5 |
6 |
0.98 |
| chr10_134266352_134266533 | 0.13 |
C10orf91 |
chromosome 10 open reading frame 91 |
7749 |
0.19 |
| chr18_43761205_43761356 | 0.13 |
C18orf25 |
chromosome 18 open reading frame 25 |
7280 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.1 | 0.2 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.1 | 0.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.1 | 0.2 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.0 | GO:0072160 | nephron tubule epithelial cell differentiation(GO:0072160) regulation of nephron tubule epithelial cell differentiation(GO:0072182) regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.1 | GO:0071816 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.1 | GO:0044091 | membrane biogenesis(GO:0044091) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.0 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.0 | GO:2000351 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:1903313 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.0 | GO:0002885 | positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.0 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.0 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.0 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0071071 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.0 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.0 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0032462 | regulation of protein homooligomerization(GO:0032462) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0072189 | ureter development(GO:0072189) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.0 | GO:0072501 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0030260 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.0 | 0.0 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.0 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0031083 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.0 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.0 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |