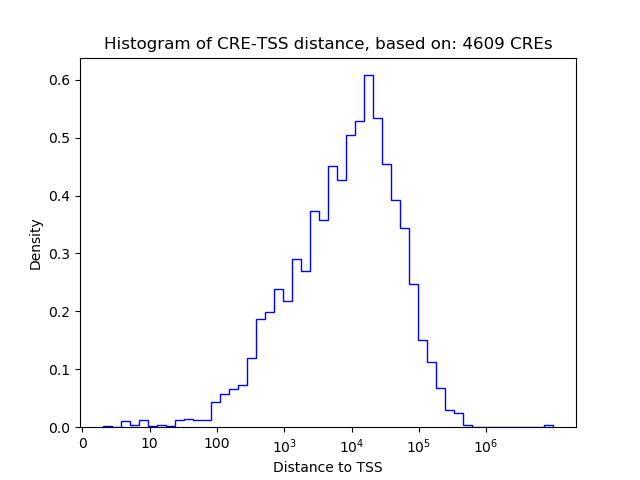
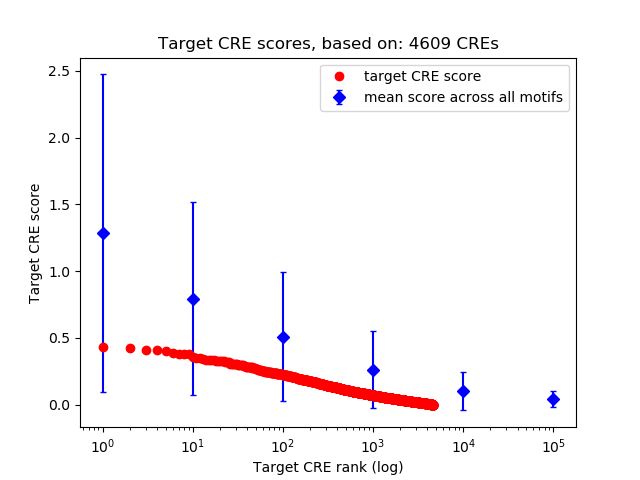
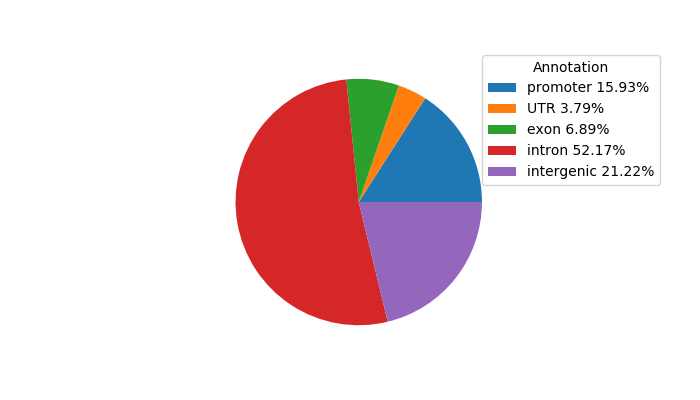
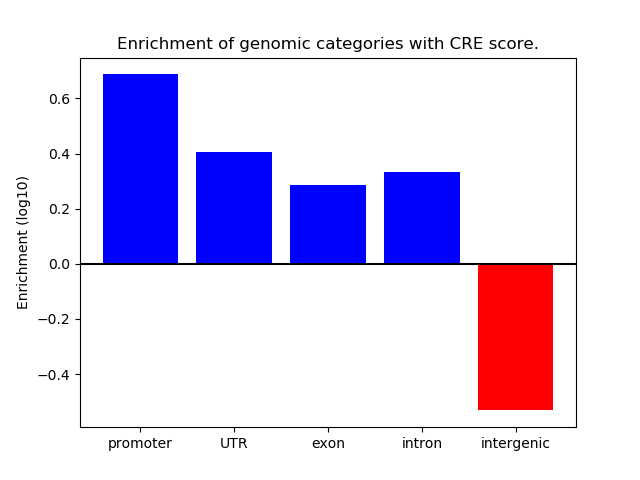
Project
ENCODE: H3K4me1 ChIP-Seq of primary human cells
Navigation
Downloads
Results for ZNF8
Z-value: 1.16
Transcription factors associated with ZNF8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF8
|
ENSG00000083842.8 | zinc finger protein 8 |
|
ZNF8
|
ENSG00000273439.1 | zinc finger protein 8 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_58790510_58790661 | ZNF8 | 267 | 0.591708 | -0.50 | 1.7e-01 | Click! |
| chr19_58792796_58792947 | ZNF8 | 2553 | 0.133213 | 0.50 | 1.8e-01 | Click! |
| chr19_58791136_58791498 | ZNF8 | 999 | 0.283145 | -0.41 | 2.7e-01 | Click! |
| chr19_58792474_58792625 | ZNF8 | 2231 | 0.145640 | 0.36 | 3.4e-01 | Click! |
| chr19_58790908_58791124 | ZNF8 | 698 | 0.373969 | -0.31 | 4.1e-01 | Click! |
Activity of the ZNF8 motif across conditions
Conditions sorted by the z-value of the ZNF8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_24867689_24867840 | 0.43 |
FAM65B |
family with sequence similarity 65, member B |
9777 |
0.19 |
| chr2_62441057_62441221 | 0.42 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
1514 |
0.37 |
| chr12_56554594_56555181 | 0.41 |
MYL6 |
myosin, light chain 6, alkali, smooth muscle and non-muscle |
2707 |
0.09 |
| chr3_49003922_49004166 | 0.41 |
RP13-131K19.1 |
|
17255 |
0.08 |
| chr22_31673029_31673483 | 0.40 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
15125 |
0.1 |
| chr12_65025056_65025207 | 0.39 |
RP11-338E21.2 |
|
3007 |
0.17 |
| chr10_29938536_29938687 | 0.38 |
SVIL |
supervillin |
14710 |
0.21 |
| chr19_57078181_57078506 | 0.38 |
AC007228.11 |
|
437 |
0.51 |
| chrX_138287334_138287495 | 0.38 |
FGF13 |
fibroblast growth factor 13 |
246 |
0.96 |
| chr13_32399322_32399473 | 0.36 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
85718 |
0.1 |
| chr21_46645058_46645209 | 0.35 |
ADARB1 |
adenosine deaminase, RNA-specific, B1 |
53576 |
0.1 |
| chr3_50231195_50231703 | 0.35 |
GNAT1 |
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
1344 |
0.29 |
| chr4_120288345_120288496 | 0.34 |
ENSG00000201186 |
. |
297 |
0.91 |
| chr18_74747142_74747293 | 0.34 |
MBP |
myelin basic protein |
17483 |
0.25 |
| chr6_35578924_35579366 | 0.34 |
ENSG00000212579 |
. |
40450 |
0.12 |
| chr7_114583964_114584237 | 0.34 |
MDFIC |
MyoD family inhibitor domain containing |
10176 |
0.32 |
| chr6_135346444_135346595 | 0.33 |
HBS1L |
HBS1-like (S. cerevisiae) |
1775 |
0.4 |
| chr5_79530041_79530192 | 0.33 |
ENSG00000239159 |
. |
5292 |
0.21 |
| chr5_149865294_149865685 | 0.32 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
108 |
0.97 |
| chr10_6597209_6597469 | 0.32 |
PRKCQ |
protein kinase C, theta |
24862 |
0.26 |
| chr4_108992806_108993006 | 0.32 |
HADH |
hydroxyacyl-CoA dehydrogenase |
67115 |
0.1 |
| chr19_41313321_41313683 | 0.32 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
354 |
0.81 |
| chr5_95992051_95992224 | 0.32 |
CAST |
calpastatin |
5640 |
0.21 |
| chr5_35828451_35828602 | 0.32 |
CTD-2113L7.1 |
|
1406 |
0.44 |
| chr8_91654470_91654621 | 0.32 |
TMEM64 |
transmembrane protein 64 |
3195 |
0.27 |
| chr18_2993997_2994477 | 0.31 |
LPIN2 |
lipin 2 |
11366 |
0.16 |
| chr13_31099418_31099569 | 0.31 |
HMGB1 |
high mobility group box 1 |
58842 |
0.14 |
| chr11_35586145_35586296 | 0.30 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
34372 |
0.18 |
| chr2_46579537_46579731 | 0.30 |
EPAS1 |
endothelial PAS domain protein 1 |
55093 |
0.13 |
| chr8_145808353_145808654 | 0.30 |
CTD-2517M22.9 |
|
921 |
0.39 |
| chr14_69252719_69253010 | 0.30 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
5096 |
0.24 |
| chr3_71265807_71265958 | 0.30 |
FOXP1 |
forkhead box P1 |
18350 |
0.27 |
| chr16_10275076_10275838 | 0.30 |
GRIN2A |
glutamate receptor, ionotropic, N-methyl D-aspartate 2A |
467 |
0.87 |
| chr4_154391718_154392045 | 0.30 |
KIAA0922 |
KIAA0922 |
4380 |
0.29 |
| chr10_101089012_101089287 | 0.30 |
CNNM1 |
cyclin M1 |
4 |
0.99 |
| chr7_150674322_150674753 | 0.29 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
477 |
0.73 |
| chr6_167364098_167364412 | 0.29 |
RP11-514O12.4 |
|
5357 |
0.16 |
| chr19_1446084_1446235 | 0.29 |
APC2 |
adenomatosis polyposis coli 2 |
141 |
0.88 |
| chr3_152101319_152101736 | 0.28 |
MBNL1 |
muscleblind-like splicing regulator 1 |
31320 |
0.18 |
| chr3_13429216_13429367 | 0.28 |
NUP210 |
nucleoporin 210kDa |
32518 |
0.2 |
| chr20_48011566_48011717 | 0.28 |
KCNB1 |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
87543 |
0.07 |
| chr19_2524704_2524855 | 0.28 |
ENSG00000252962 |
. |
21640 |
0.14 |
| chr15_31506827_31507679 | 0.28 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
53777 |
0.15 |
| chrX_78406329_78406539 | 0.28 |
GPR174 |
G protein-coupled receptor 174 |
20035 |
0.29 |
| chr1_115014648_115014799 | 0.28 |
TRIM33 |
tripartite motif containing 33 |
8675 |
0.23 |
| chr9_125570423_125570637 | 0.28 |
OR1K1 |
olfactory receptor, family 1, subfamily K, member 1 |
8160 |
0.13 |
| chr16_2077290_2077691 | 0.28 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
561 |
0.45 |
| chr7_114655426_114655724 | 0.28 |
MDFIC |
MyoD family inhibitor domain containing |
81651 |
0.11 |
| chr18_21268730_21268881 | 0.27 |
LAMA3 |
laminin, alpha 3 |
602 |
0.78 |
| chr10_48477172_48477323 | 0.27 |
GDF10 |
growth differentiation factor 10 |
38271 |
0.15 |
| chr21_15924197_15924348 | 0.27 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
5608 |
0.26 |
| chr16_58550127_58550607 | 0.27 |
SETD6 |
SET domain containing 6 |
950 |
0.45 |
| chr11_1870887_1871464 | 0.26 |
LSP1 |
lymphocyte-specific protein 1 |
3025 |
0.13 |
| chr21_32530607_32530758 | 0.26 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
28143 |
0.24 |
| chr6_45409314_45409465 | 0.26 |
RUNX2 |
runt-related transcription factor 2 |
19167 |
0.21 |
| chr2_225746186_225746337 | 0.26 |
DOCK10 |
dedicator of cytokinesis 10 |
64892 |
0.15 |
| chr3_134517935_134518086 | 0.26 |
EPHB1 |
EPH receptor B1 |
3027 |
0.27 |
| chr6_131424888_131425039 | 0.26 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
31843 |
0.21 |
| chr21_35463147_35463298 | 0.26 |
SLC5A3 |
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
17352 |
0.2 |
| chr2_204591311_204591600 | 0.26 |
CD28 |
CD28 molecule |
20039 |
0.2 |
| chr19_35670959_35671110 | 0.26 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
16096 |
0.1 |
| chr19_45999731_46000044 | 0.25 |
RTN2 |
reticulon 2 |
426 |
0.53 |
| chr16_12172780_12173025 | 0.25 |
RP11-276H1.3 |
|
11257 |
0.16 |
| chr15_78358944_78359095 | 0.25 |
TBC1D2B |
TBC1 domain family, member 2B |
554 |
0.68 |
| chr7_24324800_24324951 | 0.25 |
NPY |
neuropeptide Y |
149 |
0.96 |
| chr6_90788187_90788431 | 0.25 |
ENSG00000222078 |
. |
77084 |
0.1 |
| chr5_66452974_66453257 | 0.25 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
14853 |
0.25 |
| chr5_66816055_66816206 | 0.25 |
ENSG00000222939 |
. |
55694 |
0.18 |
| chr11_119203900_119204051 | 0.24 |
RNF26 |
ring finger protein 26 |
1262 |
0.25 |
| chr10_15183836_15183987 | 0.24 |
NMT2 |
N-myristoyltransferase 2 |
18213 |
0.17 |
| chr2_191728321_191728478 | 0.24 |
GLS |
glutaminase |
17154 |
0.2 |
| chr5_110581390_110581541 | 0.24 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
21681 |
0.19 |
| chr14_90148740_90149294 | 0.24 |
ENSG00000200312 |
. |
29838 |
0.14 |
| chr15_77575302_77575453 | 0.24 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
27483 |
0.24 |
| chr2_30457620_30458309 | 0.24 |
LBH |
limb bud and heart development |
2918 |
0.32 |
| chr4_7786856_7787007 | 0.24 |
AFAP1-AS1 |
AFAP1 antisense RNA 1 |
31114 |
0.19 |
| chrX_72783471_72783622 | 0.24 |
CHIC1 |
cysteine-rich hydrophobic domain 1 |
502 |
0.88 |
| chr19_10515778_10516368 | 0.24 |
ENSG00000221566 |
. |
1859 |
0.18 |
| chr1_227949174_227949396 | 0.24 |
SNAP47 |
synaptosomal-associated protein, 47kDa |
13521 |
0.16 |
| chr7_139313866_139314147 | 0.24 |
CLEC2L |
C-type lectin domain family 2, member L |
105178 |
0.06 |
| chr6_167524624_167524999 | 0.24 |
CCR6 |
chemokine (C-C motif) receptor 6 |
484 |
0.81 |
| chr5_38368443_38368594 | 0.24 |
EGFLAM-AS3 |
EGFLAM antisense RNA 3 |
21865 |
0.16 |
| chr18_60868113_60868539 | 0.23 |
ENSG00000238988 |
. |
6428 |
0.25 |
| chr13_74607328_74607479 | 0.23 |
KLF12 |
Kruppel-like factor 12 |
38217 |
0.23 |
| chr5_141468134_141468285 | 0.23 |
NDFIP1 |
Nedd4 family interacting protein 1 |
19861 |
0.2 |
| chr1_204495375_204495526 | 0.23 |
MDM4 |
Mdm4 p53 binding protein homolog (mouse) |
803 |
0.62 |
| chr10_14102538_14102689 | 0.23 |
RP11-397C18.2 |
|
13670 |
0.23 |
| chr5_59783822_59783973 | 0.23 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
6 |
0.99 |
| chr10_8443695_8443872 | 0.23 |
ENSG00000212505 |
. |
255011 |
0.02 |
| chr17_63175926_63176077 | 0.23 |
RGS9 |
regulator of G-protein signaling 9 |
42409 |
0.18 |
| chr15_68368372_68368523 | 0.23 |
PIAS1 |
protein inhibitor of activated STAT, 1 |
21533 |
0.24 |
| chr2_149281301_149281506 | 0.23 |
MBD5 |
methyl-CpG binding domain protein 5 |
55109 |
0.16 |
| chr10_6079510_6080013 | 0.23 |
IL2RA |
interleukin 2 receptor, alpha |
11797 |
0.14 |
| chr6_83552052_83552203 | 0.23 |
RP11-445L13__B.3 |
|
90193 |
0.1 |
| chr7_105901515_105901666 | 0.23 |
NAMPT |
nicotinamide phosphoribosyltransferase |
23777 |
0.19 |
| chr3_106055125_106055276 | 0.23 |
ENSG00000200610 |
. |
179544 |
0.03 |
| chr17_1935214_1935393 | 0.23 |
DPH1 |
diphthamide biosynthesis 1 |
1384 |
0.22 |
| chr9_135224510_135224806 | 0.23 |
SETX |
senataxin |
5714 |
0.24 |
| chr17_35030341_35030615 | 0.23 |
MRM1 |
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
72087 |
0.08 |
| chr3_197808077_197808536 | 0.23 |
ANKRD18DP |
ankyrin repeat domain 18D, pseudogene |
923 |
0.53 |
| chr15_60874078_60874257 | 0.22 |
RORA |
RAR-related orphan receptor A |
10573 |
0.22 |
| chr5_141508964_141509115 | 0.22 |
NDFIP1 |
Nedd4 family interacting protein 1 |
20969 |
0.22 |
| chr1_207503628_207503779 | 0.22 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
8567 |
0.27 |
| chr12_92949909_92950139 | 0.22 |
ENSG00000238865 |
. |
11261 |
0.26 |
| chr10_17275718_17275927 | 0.22 |
RP11-124N14.3 |
|
1010 |
0.46 |
| chr12_121845210_121845376 | 0.22 |
RNF34 |
ring finger protein 34, E3 ubiquitin protein ligase |
1965 |
0.35 |
| chr9_115868029_115868319 | 0.22 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
45048 |
0.14 |
| chr7_130577577_130577746 | 0.22 |
ENSG00000226380 |
. |
15363 |
0.25 |
| chr3_113937014_113937168 | 0.22 |
RP11-553L6.2 |
|
3853 |
0.21 |
| chr1_6505054_6505292 | 0.22 |
RP1-202O8.2 |
|
1893 |
0.19 |
| chr5_32145798_32145949 | 0.22 |
ENSG00000200065 |
. |
11029 |
0.18 |
| chr8_144753183_144753334 | 0.22 |
ZNF707 |
zinc finger protein 707 |
13364 |
0.07 |
| chr9_130318408_130318749 | 0.22 |
FAM129B |
family with sequence similarity 129, member B |
12789 |
0.17 |
| chr1_98257638_98257789 | 0.22 |
DPYD-AS2 |
DPYD antisense RNA 2 |
4764 |
0.35 |
| chr1_249116365_249116887 | 0.22 |
ENSG00000264500 |
. |
3950 |
0.17 |
| chr16_67719327_67719733 | 0.21 |
C16orf86 |
chromosome 16 open reading frame 86 |
18737 |
0.09 |
| chr15_38978663_38978814 | 0.21 |
C15orf53 |
chromosome 15 open reading frame 53 |
10061 |
0.29 |
| chr9_33141497_33141999 | 0.21 |
B4GALT1 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
13551 |
0.15 |
| chr2_38970049_38970200 | 0.21 |
SRSF7 |
serine/arginine-rich splicing factor 7 |
6582 |
0.18 |
| chr19_37998405_37998589 | 0.21 |
ZNF793 |
zinc finger protein 793 |
377 |
0.57 |
| chr17_45815718_45815869 | 0.21 |
TBX21 |
T-box 21 |
5183 |
0.16 |
| chr17_7607725_7607908 | 0.21 |
EFNB3 |
ephrin-B3 |
704 |
0.47 |
| chr3_141027177_141027447 | 0.21 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
15743 |
0.21 |
| chr5_56413254_56413405 | 0.21 |
ENSG00000207293 |
. |
51707 |
0.14 |
| chr3_15331279_15331629 | 0.21 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
13902 |
0.14 |
| chr11_126163833_126164034 | 0.21 |
RP11-712L6.7 |
|
216 |
0.87 |
| chr14_50453460_50453715 | 0.21 |
C14orf182 |
chromosome 14 open reading frame 182 |
20651 |
0.16 |
| chr10_33599602_33599753 | 0.21 |
ENSG00000244356 |
. |
22148 |
0.21 |
| chr10_33315822_33316332 | 0.21 |
ENSG00000265319 |
. |
7775 |
0.23 |
| chr3_37292881_37293032 | 0.21 |
RP11-259K5.2 |
|
7288 |
0.17 |
| chr15_39048258_39048409 | 0.21 |
C15orf53 |
chromosome 15 open reading frame 53 |
59534 |
0.15 |
| chr11_66081208_66081956 | 0.21 |
RP11-867G23.13 |
|
1258 |
0.19 |
| chr22_31335936_31336207 | 0.21 |
MORC2 |
MORC family CW-type zinc finger 2 |
7190 |
0.16 |
| chr14_56739527_56739678 | 0.21 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
153775 |
0.04 |
| chr9_123686052_123686203 | 0.20 |
TRAF1 |
TNF receptor-associated factor 1 |
4920 |
0.24 |
| chr13_95836985_95837151 | 0.20 |
ENSG00000238463 |
. |
25530 |
0.23 |
| chr17_43317813_43317964 | 0.20 |
FMNL1 |
formin-like 1 |
554 |
0.43 |
| chr8_97602268_97602419 | 0.20 |
SDC2 |
syndecan 2 |
3728 |
0.33 |
| chr20_57236648_57237104 | 0.20 |
STX16 |
syntaxin 16 |
5700 |
0.18 |
| chr1_24806575_24806726 | 0.20 |
RCAN3 |
RCAN family member 3 |
22737 |
0.15 |
| chr19_14523781_14523932 | 0.20 |
DDX39A |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A |
138 |
0.93 |
| chr22_20919945_20920204 | 0.20 |
MED15 |
mediator complex subunit 15 |
14497 |
0.14 |
| chr7_107126814_107127106 | 0.20 |
GPR22 |
G protein-coupled receptor 22 |
16455 |
0.2 |
| chr7_150263781_150263961 | 0.20 |
GIMAP4 |
GTPase, IMAP family member 4 |
494 |
0.81 |
| chr19_45264347_45264498 | 0.20 |
ENSG00000253027 |
. |
3785 |
0.13 |
| chr19_4969744_4969895 | 0.20 |
KDM4B |
lysine (K)-specific demethylase 4B |
676 |
0.68 |
| chr3_53137929_53138080 | 0.20 |
RFT1 |
RFT1 homolog (S. cerevisiae) |
26423 |
0.15 |
| chr7_130752755_130752906 | 0.19 |
ENSG00000199627 |
. |
13079 |
0.2 |
| chr12_62665115_62665307 | 0.19 |
USP15 |
ubiquitin specific peptidase 15 |
7198 |
0.22 |
| chr7_127743675_127744013 | 0.19 |
ENSG00000207588 |
. |
21931 |
0.21 |
| chr1_100877624_100878071 | 0.19 |
ENSG00000216067 |
. |
33516 |
0.16 |
| chr4_173697455_173697606 | 0.19 |
ENSG00000241652 |
. |
365264 |
0.01 |
| chr7_42882815_42882966 | 0.19 |
C7orf25 |
chromosome 7 open reading frame 25 |
68619 |
0.11 |
| chr3_13467417_13467568 | 0.19 |
NUP210 |
nucleoporin 210kDa |
5683 |
0.25 |
| chr17_29398999_29399150 | 0.19 |
RP11-271K11.5 |
|
21030 |
0.11 |
| chr9_92077290_92077553 | 0.19 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
6553 |
0.25 |
| chr4_110620562_110620713 | 0.19 |
CASP6 |
caspase 6, apoptosis-related cysteine peptidase |
3368 |
0.22 |
| chr17_41098730_41098881 | 0.19 |
ENSG00000212149 |
. |
6086 |
0.08 |
| chr2_113409796_113409947 | 0.19 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
4592 |
0.23 |
| chr3_16955552_16955703 | 0.19 |
PLCL2 |
phospholipase C-like 2 |
18955 |
0.22 |
| chr10_33444834_33444985 | 0.19 |
NRP1 |
neuropilin 1 |
29674 |
0.18 |
| chr3_30688418_30688599 | 0.19 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
40415 |
0.19 |
| chr2_39443017_39443168 | 0.19 |
CDKL4 |
cyclin-dependent kinase-like 4 |
13581 |
0.2 |
| chr2_61691635_61691786 | 0.19 |
USP34 |
ubiquitin specific peptidase 34 |
6194 |
0.19 |
| chr15_50722930_50723081 | 0.19 |
USP8 |
ubiquitin specific peptidase 8 |
3914 |
0.2 |
| chr4_109348290_109348441 | 0.19 |
ENSG00000266046 |
. |
149931 |
0.04 |
| chr9_89885412_89885563 | 0.19 |
ENSG00000212421 |
. |
10122 |
0.3 |
| chr16_67226498_67226740 | 0.19 |
E2F4 |
E2F transcription factor 4, p107/p130-binding |
547 |
0.48 |
| chr10_104169426_104169807 | 0.19 |
PSD |
pleckstrin and Sec7 domain containing |
9285 |
0.1 |
| chr16_87695421_87695572 | 0.19 |
AC010536.1 |
Uncharacterized protein; cDNA FLJ45526 fis, clone BRTHA2027227 |
34257 |
0.13 |
| chr16_29713724_29714191 | 0.19 |
QPRT |
quinolinate phosphoribosyltransferase |
23454 |
0.09 |
| chr8_127287160_127287311 | 0.19 |
ENSG00000221706 |
. |
133232 |
0.05 |
| chr12_50138993_50139144 | 0.18 |
TMBIM6 |
transmembrane BAX inhibitor motif containing 6 |
1868 |
0.31 |
| chr2_235321270_235321441 | 0.18 |
ARL4C |
ADP-ribosylation factor-like 4C |
83889 |
0.11 |
| chr7_16968826_16969134 | 0.18 |
AGR3 |
anterior gradient 3 |
47369 |
0.17 |
| chr17_45899312_45899856 | 0.18 |
OSBPL7 |
oxysterol binding protein-like 7 |
384 |
0.77 |
| chr13_51523103_51523254 | 0.18 |
RNASEH2B-AS1 |
RNASEH2B antisense RNA 1 |
38330 |
0.15 |
| chrX_48583462_48583613 | 0.18 |
ENSG00000206723 |
. |
571 |
0.62 |
| chr8_57069675_57070078 | 0.18 |
ENSG00000221093 |
. |
35965 |
0.12 |
| chr19_17597784_17597935 | 0.18 |
SLC27A1 |
solute carrier family 27 (fatty acid transporter), member 1 |
13316 |
0.08 |
| chr3_38691341_38691875 | 0.18 |
SCN5A |
sodium channel, voltage-gated, type V, alpha subunit |
444 |
0.88 |
| chr2_106702316_106702467 | 0.18 |
C2orf40 |
chromosome 2 open reading frame 40 |
20135 |
0.23 |
| chrX_48584106_48584257 | 0.18 |
ENSG00000206723 |
. |
1215 |
0.32 |
| chr14_51708170_51708339 | 0.18 |
TMX1 |
thioredoxin-related transmembrane protein 1 |
1368 |
0.39 |
| chr11_128499546_128499697 | 0.18 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
42168 |
0.14 |
| chr11_62489486_62489689 | 0.18 |
HNRNPUL2 |
heterogeneous nuclear ribonucleoprotein U-like 2 |
5234 |
0.07 |
| chr5_98114835_98114986 | 0.18 |
RGMB |
repulsive guidance molecule family member b |
5571 |
0.24 |
| chr9_92151633_92152777 | 0.18 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
39160 |
0.16 |
| chr7_156746508_156746659 | 0.18 |
NOM1 |
nucleolar protein with MIF4G domain 1 |
4166 |
0.22 |
| chr4_95133944_95134095 | 0.18 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
4821 |
0.35 |
| chr17_37922924_37923075 | 0.18 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
11479 |
0.12 |
| chr2_102439045_102439294 | 0.18 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
17121 |
0.26 |
| chrX_23684782_23684975 | 0.18 |
PRDX4 |
peroxiredoxin 4 |
685 |
0.75 |
| chr4_147576742_147576893 | 0.18 |
ENSG00000264323 |
. |
16404 |
0.21 |
| chr1_219348883_219349034 | 0.18 |
RP11-135J2.4 |
|
1655 |
0.37 |
| chr19_39835822_39835973 | 0.18 |
SAMD4B |
sterile alpha motif domain containing 4B |
2789 |
0.12 |
| chr11_118276298_118276449 | 0.18 |
RP11-770J1.5 |
Uncharacterized protein |
3763 |
0.12 |
| chr16_67338311_67338462 | 0.18 |
KCTD19 |
potassium channel tetramerization domain containing 19 |
10660 |
0.09 |
| chr21_34920280_34920507 | 0.18 |
SON |
SON DNA binding protein |
4161 |
0.15 |
| chr12_777327_777560 | 0.18 |
NINJ2 |
ninjurin 2 |
4498 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.3 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 0.2 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0002517 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.0 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.2 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0034776 | response to histamine(GO:0034776) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:1903312 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.0 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.0 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0090312 | regulation of protein deacetylation(GO:0090311) positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.0 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.0 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0042517 | positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.0 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.0 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.0 | GO:1904035 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0061630 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |