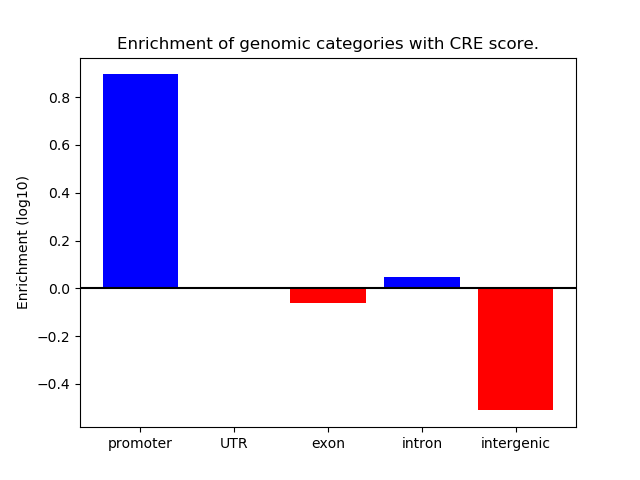
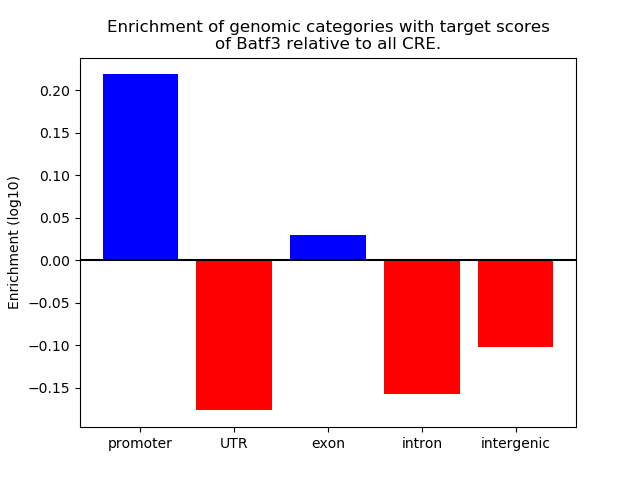
Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
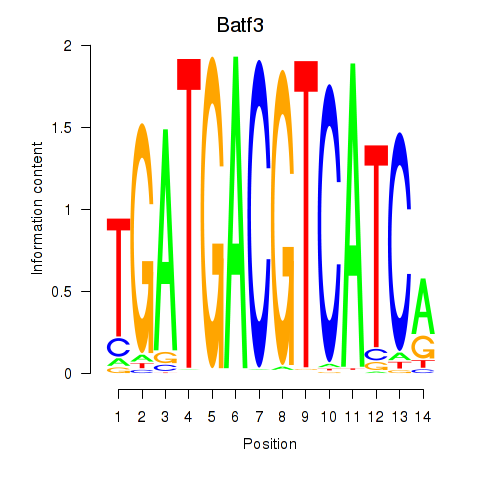
Results for Batf3
Z-value: 0.62
Transcription factors associated with Batf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Batf3
|
ENSMUSG00000026630.4 | basic leucine zipper transcription factor, ATF-like 3 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_191097955_191098106 | Batf3 | 150 | 0.924944 | -0.28 | 3.9e-02 | Click! |
| chr1_191098208_191098740 | Batf3 | 91 | 0.947370 | -0.23 | 8.9e-02 | Click! |
| chr1_191111292_191111443 | Batf3 | 10890 | 0.113908 | -0.14 | 3.0e-01 | Click! |
Activity of the Batf3 motif across conditions
Conditions sorted by the z-value of the Batf3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
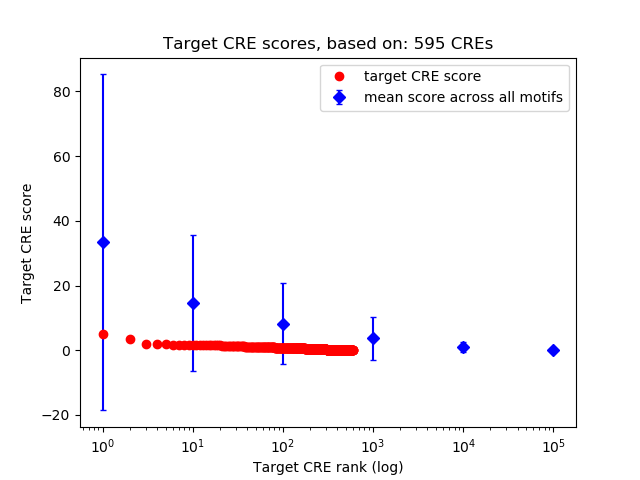
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_69709128_69709981 | 4.91 |
Rpl32-ps |
ribosomal protein L32, pseudogene |
7839 |
0.17 |
| chr1_165396068_165396234 | 3.55 |
Dcaf6 |
DDB1 and CUL4 associated factor 6 |
7390 |
0.14 |
| chr2_49730225_49730511 | 1.99 |
Kif5c |
kinesin family member 5C |
2243 |
0.36 |
| chr3_17793443_17793892 | 1.96 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
253 |
0.9 |
| chr7_142088957_142089418 | 1.92 |
Dusp8 |
dual specificity phosphatase 8 |
6085 |
0.08 |
| chr17_72399498_72399651 | 1.76 |
Gm24736 |
predicted gene, 24736 |
99844 |
0.07 |
| chr6_124464616_124464917 | 1.75 |
Clstn3 |
calsyntenin 3 |
28 |
0.95 |
| chr1_38446167_38446442 | 1.74 |
Gm34727 |
predicted gene, 34727 |
41215 |
0.16 |
| chr3_129536267_129536526 | 1.72 |
Gm43072 |
predicted gene 43072 |
1745 |
0.29 |
| chr8_70500802_70501205 | 1.68 |
Crlf1 |
cytokine receptor-like factor 1 |
130 |
0.9 |
| chr14_68048454_68048605 | 1.67 |
Gm31107 |
predicted gene, 31107 |
2801 |
0.28 |
| chr12_73286641_73287386 | 1.64 |
Slc38a6 |
solute carrier family 38, member 6 |
47 |
0.81 |
| chr14_87140676_87141090 | 1.61 |
Diaph3 |
diaphanous related formin 3 |
260 |
0.93 |
| chr6_57703265_57703437 | 1.60 |
Lancl2 |
LanC (bacterial lantibiotic synthetase component C)-like 2 |
264 |
0.88 |
| chr7_141146101_141146426 | 1.60 |
Ptdss2 |
phosphatidylserine synthase 2 |
8360 |
0.08 |
| chr13_65258166_65258818 | 1.59 |
Gm10775 |
predicted gene 10775 |
1047 |
0.28 |
| chr8_60954723_60954914 | 1.55 |
Clcn3 |
chloride channel, voltage-sensitive 3 |
39 |
0.97 |
| chr13_22042530_22043117 | 1.54 |
H2ac11 |
H2A clustered histone 11 |
121 |
0.56 |
| chr6_114659319_114659576 | 1.47 |
Atg7 |
autophagy related 7 |
1383 |
0.51 |
| chr7_97749679_97749846 | 1.46 |
Aqp11 |
aquaporin 11 |
11473 |
0.16 |
| chr4_142017787_142018051 | 1.42 |
4930455G09Rik |
RIKEN cDNA 4930455G09 gene |
21 |
0.96 |
| chr14_74972805_74972992 | 1.42 |
Gm4278 |
predicted gene 4278 |
2188 |
0.3 |
| chr7_128414880_128415076 | 1.40 |
Gm15503 |
predicted gene 15503 |
2637 |
0.17 |
| chr2_151563228_151563425 | 1.37 |
Sdcbp2 |
syndecan binding protein (syntenin) 2 |
9296 |
0.13 |
| chr11_103101909_103102424 | 1.37 |
Acbd4 |
acyl-Coenzyme A binding domain containing 4 |
440 |
0.48 |
| chrY_90771156_90771656 | 1.35 |
Gm47283 |
predicted gene, 47283 |
13332 |
0.16 |
| chr14_61680824_61681965 | 1.35 |
Gm37472 |
predicted gene, 37472 |
100 |
0.9 |
| chrX_169996827_169997318 | 1.34 |
Gm15247 |
predicted gene 15247 |
10133 |
0.15 |
| chr1_82836684_82836853 | 1.30 |
Gm22396 |
predicted gene, 22396 |
2678 |
0.14 |
| chr9_111534874_111535371 | 1.30 |
Gm42523 |
predicted gene 42523 |
13818 |
0.19 |
| chr3_96905206_96905674 | 1.28 |
Gpr89 |
G protein-coupled receptor 89 |
94 |
0.91 |
| chr12_3237774_3237992 | 1.26 |
Rab10os |
RAB10, member RAS oncogene family, opposite strand |
1272 |
0.39 |
| chr17_55892287_55892468 | 1.21 |
Zfp959 |
zinc finger protein 959 |
91 |
0.95 |
| chr15_27871362_27871539 | 1.18 |
Gm20555 |
predicted gene, 20555 |
4219 |
0.22 |
| chr17_55878572_55878762 | 1.18 |
Zfp119a |
zinc finger protein 119a |
263 |
0.86 |
| chr6_120807499_120807674 | 1.17 |
Atp6v1e1 |
ATPase, H+ transporting, lysosomal V1 subunit E1 |
5164 |
0.15 |
| chr18_44828099_44828778 | 1.15 |
Ythdc2 |
YTH domain containing 2 |
692 |
0.64 |
| chr9_123925253_123925437 | 1.13 |
Ccr1 |
chemokine (C-C motif) receptor 1 |
43347 |
0.1 |
| chr1_132322162_132322332 | 1.13 |
Nuak2 |
NUAK family, SNF1-like kinase, 2 |
5349 |
0.13 |
| chr1_179433930_179434100 | 1.12 |
Smyd3 |
SET and MYND domain containing 3 |
28596 |
0.2 |
| chr12_3238767_3239202 | 1.11 |
Rab10os |
RAB10, member RAS oncogene family, opposite strand |
2373 |
0.24 |
| chr10_63122610_63122870 | 1.11 |
Mypn |
myopalladin |
56 |
0.96 |
| chr3_17592524_17592749 | 1.10 |
Gm38154 |
predicted gene, 38154 |
78052 |
0.11 |
| chr2_129227265_129227416 | 1.10 |
A730036I17Rik |
RIKEN cDNA A730036I17 gene |
682 |
0.4 |
| chr6_115666689_115667030 | 1.10 |
Raf1 |
v-raf-leukemia viral oncogene 1 |
9105 |
0.12 |
| chr13_23760830_23761749 | 1.09 |
H4c1 |
H4 clustered histone 1 |
59 |
0.86 |
| chr8_79711585_79712295 | 1.07 |
Anapc10 |
anaphase promoting complex subunit 10 |
89 |
0.65 |
| chr16_75837887_75838038 | 1.05 |
Gm15554 |
predicted gene 15554 |
64905 |
0.09 |
| chr8_77548898_77549532 | 1.05 |
Prmt9 |
protein arginine methyltransferase 9 |
182 |
0.92 |
| chr1_184851887_184852286 | 1.04 |
Mtarc2 |
mitochondrial amidoxime reducing component 2 |
5635 |
0.17 |
| chr2_59351695_59352106 | 1.04 |
Pkp4 |
plakophilin 4 |
9703 |
0.19 |
| chr7_118463178_118463349 | 1.03 |
Syt17 |
synaptotagmin XVII |
15041 |
0.15 |
| chr11_88242035_88242186 | 1.03 |
Gm38534 |
predicted gene, 38534 |
11258 |
0.17 |
| chr3_94484140_94484361 | 1.02 |
Celf3 |
CUGBP, Elav-like family member 3 |
77 |
0.93 |
| chr2_109280292_109280860 | 1.01 |
Mettl15 |
methyltransferase like 15 |
126 |
0.55 |
| chr16_10976223_10976631 | 1.00 |
Litaf |
LPS-induced TN factor |
848 |
0.46 |
| chr6_128799495_128800104 | 1.00 |
Klrb1c |
killer cell lectin-like receptor subfamily B member 1C |
10584 |
0.1 |
| chr1_75214435_75214934 | 0.98 |
Stk16 |
serine/threonine kinase 16 |
1623 |
0.13 |
| chr6_57822566_57822717 | 0.97 |
Vopp1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
1999 |
0.23 |
| chr12_84217180_84217402 | 0.96 |
Gm47447 |
predicted gene, 47447 |
1154 |
0.3 |
| chr2_127444443_127444898 | 0.96 |
Fahd2a |
fumarylacetoacetate hydrolase domain containing 2A |
105 |
0.96 |
| chr9_103234179_103234348 | 0.95 |
Trf |
transferrin |
3819 |
0.2 |
| chr3_82478392_82478716 | 0.95 |
Npy2r |
neuropeptide Y receptor Y2 |
69406 |
0.11 |
| chr7_97788183_97788962 | 0.94 |
Pak1 |
p21 (RAC1) activated kinase 1 |
31 |
0.98 |
| chr12_100167000_100167151 | 0.93 |
Nrde2 |
nrde-2 necessary for RNA interference, domain containing |
7422 |
0.14 |
| chr17_55944886_55945074 | 0.93 |
Zfp119b |
zinc finger protein 119b |
279 |
0.8 |
| chr8_66860040_66861058 | 0.90 |
Naf1 |
nuclear assembly factor 1 ribonucleoprotein |
332 |
0.88 |
| chr9_7836413_7837486 | 0.90 |
Birc2 |
baculoviral IAP repeat-containing 2 |
108 |
0.96 |
| chr13_21935906_21936057 | 0.90 |
Zfp184 |
zinc finger protein 184 (Kruppel-like) |
9113 |
0.06 |
| chrX_13070554_13071574 | 0.90 |
Usp9x |
ubiquitin specific peptidase 9, X chromosome |
434 |
0.84 |
| chr10_108333682_108333833 | 0.89 |
Pawr |
PRKC, apoptosis, WT1, regulator |
940 |
0.6 |
| chr6_86669349_86669907 | 0.88 |
Mxd1 |
MAX dimerization protein 1 |
467 |
0.68 |
| chr18_10939016_10939167 | 0.88 |
Gm7575 |
predicted gene 7575 |
10207 |
0.23 |
| chr18_9957690_9958213 | 0.88 |
Thoc1 |
THO complex 1 |
45 |
0.97 |
| chr11_4250427_4250602 | 0.87 |
Lif |
leukemia inhibitory factor |
7043 |
0.1 |
| chr13_113168382_113168574 | 0.87 |
Gzmk |
granzyme K |
12419 |
0.12 |
| chr7_80324088_80324301 | 0.85 |
Rccd1 |
RCC1 domain containing 1 |
41 |
0.95 |
| chr16_57606439_57606962 | 0.84 |
Cmss1 |
cms small ribosomal subunit 1 |
76 |
0.98 |
| chr7_138846157_138846363 | 0.84 |
Mapk1ip1 |
mitogen-activated protein kinase 1 interacting protein 1 |
0 |
0.58 |
| chr4_63374024_63374199 | 0.84 |
Akna |
AT-hook transcription factor |
7092 |
0.11 |
| chr2_93096177_93096364 | 0.83 |
Gm13802 |
predicted gene 13802 |
1974 |
0.37 |
| chr9_69467648_69467967 | 0.82 |
Anxa2 |
annexin A2 |
10658 |
0.12 |
| chr7_16296088_16297248 | 0.82 |
Ccdc9 |
coiled-coil domain containing 9 |
9873 |
0.11 |
| chr9_64178981_64179377 | 0.81 |
Snapc5 |
small nuclear RNA activating complex, polypeptide 5 |
95 |
0.89 |
| chr6_35173300_35173451 | 0.79 |
Nup205 |
nucleoporin 205 |
4046 |
0.24 |
| chr11_20200843_20201200 | 0.79 |
Rab1a |
RAB1A, member RAS oncogene family |
411 |
0.82 |
| chr3_40799878_40800077 | 0.79 |
Plk4 |
polo like kinase 4 |
42 |
0.96 |
| chr17_88113373_88113524 | 0.78 |
Gm4832 |
predicted gene 4832 |
12064 |
0.18 |
| chr17_28013289_28013809 | 0.77 |
Anks1 |
ankyrin repeat and SAM domain containing 1 |
6204 |
0.13 |
| chr1_92473261_92473440 | 0.77 |
Ndufa10 |
NADH:ubiquinone oxidoreductase subunit A10 |
397 |
0.76 |
| chr17_53596429_53596797 | 0.76 |
Gm6919 |
predicted gene 6919 |
2050 |
0.27 |
| chr4_63389695_63390110 | 0.75 |
Aknaos |
AT-hook transcription factor, opposite strand |
8544 |
0.12 |
| chr4_116175694_116176193 | 0.75 |
Gm12951 |
predicted gene 12951 |
303 |
0.78 |
| chr8_4779223_4779832 | 0.74 |
Shcbp1 |
Shc SH2-domain binding protein 1 |
24 |
0.95 |
| chr3_105483711_105484059 | 0.74 |
Gm43847 |
predicted gene 43847 |
19022 |
0.17 |
| chr2_129242545_129242834 | 0.74 |
Gm14024 |
predicted gene 14024 |
3687 |
0.12 |
| chr7_16399506_16399684 | 0.74 |
Zc3h4 |
zinc finger CCCH-type containing 4 |
1315 |
0.27 |
| chr14_30876254_30876405 | 0.74 |
Mustn1 |
musculoskeletal, embryonic nuclear protein 1 |
2871 |
0.16 |
| chrX_56597885_56598283 | 0.73 |
Mmgt1 |
membrane magnesium transporter 1 |
15 |
0.97 |
| chr5_130706297_130706454 | 0.73 |
Gm23761 |
predicted gene, 23761 |
76326 |
0.1 |
| chr1_161766742_161767051 | 0.73 |
Gm5049 |
predicted gene 5049 |
20840 |
0.12 |
| chr2_120609238_120609778 | 0.73 |
Lrrc57 |
leucine rich repeat containing 57 |
0 |
0.56 |
| chr19_61227192_61227555 | 0.73 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
3 |
0.96 |
| chr1_75168558_75168746 | 0.73 |
Zfand2b |
zinc finger, AN1 type domain 2B |
3 |
0.93 |
| chr11_121086446_121086616 | 0.72 |
Sectm1a |
secreted and transmembrane 1A |
5311 |
0.09 |
| chr19_24961254_24961969 | 0.72 |
Cbwd1 |
COBW domain containing 1 |
1 |
0.97 |
| chr18_12786714_12786865 | 0.71 |
Osbpl1a |
oxysterol binding protein-like 1A |
19652 |
0.15 |
| chr7_99795341_99796009 | 0.71 |
F730035P03Rik |
RIKEN cDNA F730035P03 gene |
14136 |
0.11 |
| chr6_49263929_49264218 | 0.71 |
Tra2a |
transformer 2 alpha |
40 |
0.97 |
| chr3_34231745_34231896 | 0.70 |
Sox2ot |
SOX2 overlapping transcript (non-protein coding) |
34956 |
0.16 |
| chr10_59777356_59777560 | 0.70 |
Micu1 |
mitochondrial calcium uptake 1 |
15045 |
0.14 |
| chr13_67360105_67360768 | 0.70 |
Gm28044 |
predicted gene, 28044 |
100 |
0.47 |
| chr13_97775152_97775346 | 0.69 |
Rps18-ps6 |
ribosomal protein S18, pseudogene 6 |
14617 |
0.14 |
| chr1_92473653_92474522 | 0.69 |
Ndufa10 |
NADH:ubiquinone oxidoreductase subunit A10 |
227 |
0.88 |
| chr12_75307963_75308582 | 0.69 |
Rhoj |
ras homolog family member J |
50 |
0.99 |
| chr7_7298676_7299288 | 0.69 |
Mir5620 |
microRNA 5620 |
91 |
0.61 |
| chr5_123854595_123854839 | 0.68 |
Hcar2 |
hydroxycarboxylic acid receptor 2 |
10782 |
0.13 |
| chr10_111073345_111073645 | 0.68 |
Gm48851 |
predicted gene, 48851 |
29376 |
0.14 |
| chr4_41708277_41708428 | 0.68 |
Rpp25l |
ribonuclease P/MRP 25 subunit-like |
5182 |
0.1 |
| chr8_84256509_84256688 | 0.68 |
Mri1 |
methylthioribose-1-phosphate isomerase 1 |
185 |
0.87 |
| chr10_82240724_82241110 | 0.67 |
Zfp938 |
zinc finger protein 938 |
356 |
0.84 |
| chr16_38562668_38563355 | 0.66 |
Tmem39a |
transmembrane protein 39a |
125 |
0.95 |
| chr9_71976147_71976576 | 0.66 |
Gm37663 |
predicted gene, 37663 |
14910 |
0.1 |
| chr9_14246488_14246694 | 0.66 |
Gm47565 |
predicted gene, 47565 |
4348 |
0.18 |
| chr7_135813684_135813871 | 0.66 |
6330420H09Rik |
RIKEN cDNA 6330420H09 gene |
39905 |
0.12 |
| chr5_89763970_89764289 | 0.66 |
Gm25758 |
predicted gene, 25758 |
64742 |
0.13 |
| chr2_20944134_20944558 | 0.66 |
Arhgap21 |
Rho GTPase activating protein 21 |
927 |
0.58 |
| chr2_69722689_69722921 | 0.66 |
Ppig |
peptidyl-prolyl isomerase G (cyclophilin G) |
0 |
0.96 |
| chr5_24383104_24384088 | 0.66 |
Gm15587 |
predicted gene 15587 |
14 |
0.94 |
| chr15_13171596_13171768 | 0.66 |
Cdh6 |
cadherin 6 |
1993 |
0.47 |
| chrX_101290140_101290457 | 0.65 |
Med12 |
mediator complex subunit 12 |
2836 |
0.13 |
| chr1_161036007_161036172 | 0.65 |
Gm22357 |
predicted gene, 22357 |
32 |
0.41 |
| chr11_29607584_29607776 | 0.65 |
Gm12091 |
predicted gene 12091 |
5067 |
0.14 |
| chr4_154603637_154604075 | 0.64 |
Gm13134 |
predicted gene 13134 |
2541 |
0.22 |
| chr17_57098452_57098603 | 0.63 |
Tnfsf9 |
tumor necrosis factor (ligand) superfamily, member 9 |
6798 |
0.08 |
| chr12_86883989_86885134 | 0.63 |
Irf2bpl |
interferon regulatory factor 2 binding protein-like |
237 |
0.93 |
| chr2_180892991_180893395 | 0.63 |
Mir124a-3 |
microRNA 124a-3 |
847 |
0.31 |
| chr2_122380827_122380978 | 0.63 |
Gm24409 |
predicted gene, 24409 |
4738 |
0.15 |
| chr9_106170484_106171031 | 0.62 |
Wdr82 |
WD repeat domain containing 82 |
171 |
0.89 |
| chr2_15054565_15055405 | 0.62 |
Nsun6 |
NOL1/NOP2/Sun domain family member 6 |
84 |
0.72 |
| chr6_21949203_21949798 | 0.61 |
Ing3 |
inhibitor of growth family, member 3 |
71 |
0.98 |
| chr7_7299356_7299778 | 0.61 |
Clcn4 |
chloride channel, voltage-sensitive 4 |
10 |
0.91 |
| chr7_4149205_4149990 | 0.61 |
Leng9 |
leukocyte receptor cluster (LRC) member 9 |
777 |
0.41 |
| chr2_125122926_125123420 | 0.61 |
Myef2 |
myelin basic protein expression factor 2, repressor |
241 |
0.89 |
| chr2_112454615_112455124 | 0.61 |
Emc7 |
ER membrane protein complex subunit 7 |
128 |
0.94 |
| chr17_84877971_84878140 | 0.60 |
Gm49982 |
predicted gene, 49982 |
24514 |
0.14 |
| chr15_7137719_7137966 | 0.60 |
Lifr |
LIF receptor alpha |
2700 |
0.38 |
| chr2_144343181_144343332 | 0.60 |
Ovol2 |
ovo like zinc finger 2 |
11110 |
0.1 |
| chr13_67724281_67724474 | 0.60 |
Zfp65 |
zinc finger protein 65 |
51 |
0.92 |
| chr1_134454955_134455620 | 0.60 |
Gm37935 |
predicted gene, 37935 |
186 |
0.56 |
| chr6_91440817_91441099 | 0.60 |
1810044D09Rik |
RIKEN cDNA 1810044D09 gene |
29 |
0.5 |
| chr4_156054432_156054595 | 0.59 |
Mir200a |
microRNA 200a |
472 |
0.37 |
| chr10_126749213_126749376 | 0.59 |
Gm47966 |
predicted gene, 47966 |
29900 |
0.12 |
| chr18_66458401_66458670 | 0.59 |
Pmaip1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
2 |
0.96 |
| chr8_69625175_69625427 | 0.59 |
Zfp868 |
zinc finger protein 868 |
244 |
0.91 |
| chrX_8176094_8176317 | 0.59 |
Tbc1d25 |
TBC1 domain family, member 25 |
24 |
0.95 |
| chr1_59237629_59237969 | 0.58 |
Als2 |
alsin Rho guanine nucleotide exchange factor |
568 |
0.74 |
| chr2_120153745_120155075 | 0.58 |
Ehd4 |
EH-domain containing 4 |
52 |
0.97 |
| chr1_166409714_166410315 | 0.57 |
Pogk |
pogo transposable element with KRAB domain |
151 |
0.94 |
| chr16_50046157_50046358 | 0.57 |
Gm8824 |
predicted gene 8824 |
64816 |
0.14 |
| chr11_116572806_116573131 | 0.57 |
Ube2o |
ubiquitin-conjugating enzyme E2O |
8479 |
0.1 |
| chr3_104795762_104796004 | 0.57 |
Rhoc |
ras homolog family member C |
3900 |
0.11 |
| chr17_63784808_63784959 | 0.57 |
Fer |
fer (fms/fps related) protein kinase |
78179 |
0.09 |
| chr19_43898677_43898828 | 0.57 |
Dnmbp |
dynamin binding protein |
8061 |
0.14 |
| chr3_127552930_127553443 | 0.56 |
Larp7 |
La ribonucleoprotein domain family, member 7 |
11 |
0.79 |
| chr2_122377386_122377671 | 0.56 |
Gm24409 |
predicted gene, 24409 |
8112 |
0.13 |
| chr10_44268190_44268842 | 0.56 |
Atg5 |
autophagy related 5 |
101 |
0.97 |
| chr4_46041817_46041999 | 0.56 |
Tmod1 |
tropomodulin 1 |
2699 |
0.27 |
| chr19_61228253_61228788 | 0.55 |
Csf2ra |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
91 |
0.95 |
| chr1_137900910_137901218 | 0.55 |
Gm4258 |
predicted gene 4258 |
2466 |
0.14 |
| chrX_155216720_155216871 | 0.55 |
Sat1 |
spermidine/spermine N1-acetyl transferase 1 |
346 |
0.82 |
| chr1_171375937_171376095 | 0.54 |
Nectin4 |
nectin cell adhesion molecule 4 |
5661 |
0.07 |
| chr2_30929577_30930014 | 0.54 |
Ptges |
prostaglandin E synthase |
68 |
0.96 |
| chr5_32196023_32196194 | 0.54 |
Gm9555 |
predicted gene 9555 |
10611 |
0.15 |
| chr18_80151657_80151990 | 0.53 |
Adnp2 |
ADNP homeobox 2 |
341 |
0.77 |
| chr13_23530854_23531458 | 0.53 |
H4c8 |
H4 clustered histone 8 |
106 |
0.6 |
| chr5_36691557_36691708 | 0.51 |
D5Ertd579e |
DNA segment, Chr 5, ERATO Doi 579, expressed |
4305 |
0.16 |
| chr5_33018563_33018867 | 0.51 |
Ywhah |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
101 |
0.96 |
| chrX_16618200_16618385 | 0.51 |
Maoa |
monoamine oxidase A |
1406 |
0.48 |
| chr10_82354457_82354651 | 0.51 |
Gm4924 |
predicted gene 4924 |
238 |
0.92 |
| chr7_27675038_27675234 | 0.51 |
Map3k10 |
mitogen-activated protein kinase kinase kinase 10 |
538 |
0.6 |
| chr1_59394147_59394298 | 0.51 |
Gm29016 |
predicted gene 29016 |
15292 |
0.18 |
| chr4_89310510_89311260 | 0.49 |
Cdkn2b |
cyclin dependent kinase inhibitor 2B |
154 |
0.95 |
| chr9_37177686_37177862 | 0.49 |
Gm25273 |
predicted gene, 25273 |
4780 |
0.14 |
| chr14_62446475_62446626 | 0.48 |
Gucy1b2 |
guanylate cyclase 1, soluble, beta 2 |
8243 |
0.18 |
| chr13_55425993_55426144 | 0.48 |
F12 |
coagulation factor XII (Hageman factor) |
711 |
0.46 |
| chr17_84466117_84466668 | 0.48 |
Thada |
thyroid adenoma associated |
187 |
0.95 |
| chrX_162902264_162902478 | 0.47 |
Ctps2 |
cytidine 5'-triphosphate synthase 2 |
200 |
0.91 |
| chr12_82225804_82226178 | 0.45 |
Sipa1l1 |
signal-induced proliferation-associated 1 like 1 |
30594 |
0.19 |
| chr14_77034576_77034752 | 0.44 |
Lacc1 |
laccase domain containing 1 |
1417 |
0.39 |
| chr2_153161013_153161878 | 0.43 |
Tm9sf4 |
transmembrane 9 superfamily protein member 4 |
96 |
0.96 |
| chr14_50807340_50808013 | 0.43 |
Ccnb1ip1 |
cyclin B1 interacting protein 1 |
56 |
0.42 |
| chr11_116657908_116658166 | 0.43 |
Prcd |
photoreceptor disc component |
881 |
0.36 |
| chr13_83737818_83738360 | 0.42 |
Gm33366 |
predicted gene, 33366 |
446 |
0.49 |
| chr10_81602742_81603198 | 0.42 |
Tle6 |
transducin-like enhancer of split 6 |
1897 |
0.15 |
| chr1_75210411_75211182 | 0.42 |
Glb1l |
galactosidase, beta 1-like |
17 |
0.49 |
| chr12_98628022_98628216 | 0.42 |
Spata7 |
spermatogenesis associated 7 |
38 |
0.96 |
| chr13_100871561_100871718 | 0.41 |
Gm37830 |
predicted gene, 37830 |
1141 |
0.42 |
| chr6_108706138_108706289 | 0.41 |
Bhlhe40 |
basic helix-loop-helix family, member e40 |
43167 |
0.12 |
| chrX_104293744_104293895 | 0.41 |
Vcp-rs |
valosin containing protein, related sequence |
54415 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.5 | 1.4 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.3 | 1.7 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.3 | 1.3 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.3 | 0.9 | GO:0048290 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.3 | 0.8 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.3 | 1.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 0.7 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 | 0.6 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.2 | 0.7 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.2 | 0.7 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.2 | 0.8 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 0.8 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 0.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.4 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.7 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.1 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.7 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.8 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.3 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.7 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.3 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.4 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.3 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.8 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.2 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.1 | 0.2 | GO:2000019 | negative regulation of male gonad development(GO:2000019) |
| 0.1 | 0.3 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.3 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.1 | 0.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.2 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.1 | 0.2 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.4 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.5 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.4 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.8 | GO:1904872 | regulation of telomerase RNA localization to Cajal body(GO:1904872) positive regulation of telomerase RNA localization to Cajal body(GO:1904874) |
| 0.1 | 0.2 | GO:1901642 | purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.2 | GO:0097460 | ferrous iron import(GO:0070627) ferrous iron import into cell(GO:0097460) |
| 0.1 | 0.2 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) |
| 0.1 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.0 | 0.5 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.2 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.1 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0010963 | regulation of L-arginine import(GO:0010963) |
| 0.0 | 1.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.6 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.1 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.5 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.6 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.5 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 1.0 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.1 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.5 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.4 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.5 | GO:0021889 | olfactory bulb interneuron differentiation(GO:0021889) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.0 | GO:0044805 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.2 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.0 | GO:0070350 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.0 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.0 | GO:0071672 | negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.0 | 0.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.3 | GO:0071549 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.4 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.0 | 0.3 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.0 | GO:1902564 | negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0042435 | indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 | 0.0 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.0 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.0 | 0.0 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 1.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.0 | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process(GO:0009219) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.0 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.0 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.0 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.0 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.8 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.0 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.0 | 0.0 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 1.0 | GO:0008213 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.1 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) |
| 0.0 | 0.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 0.5 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 0.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.9 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.3 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.8 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 1.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 0.7 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.2 | 0.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 2.0 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 1.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.4 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.8 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.6 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.1 | 0.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.8 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.2 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.5 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.9 | GO:0052770 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 2.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 1.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.0 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.1 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.0 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 1.0 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.0 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0015232 | heme transporter activity(GO:0015232) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.8 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.8 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |