Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
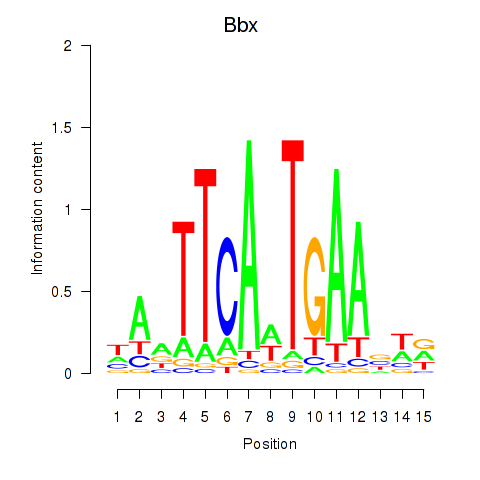
Results for Bbx
Z-value: 0.86
Transcription factors associated with Bbx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bbx
|
ENSMUSG00000022641.9 | bobby sox HMG box containing |
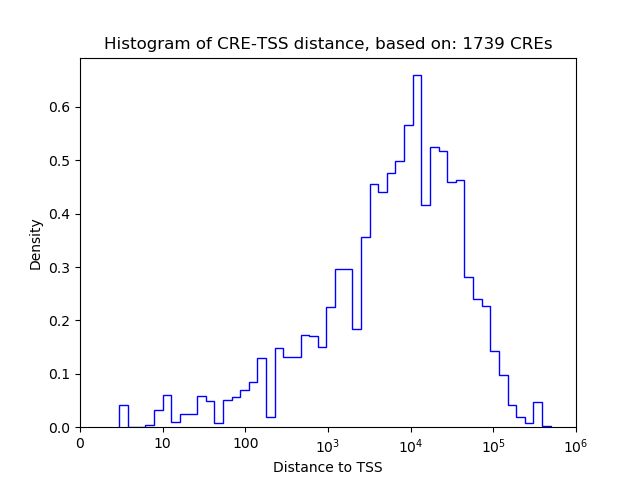
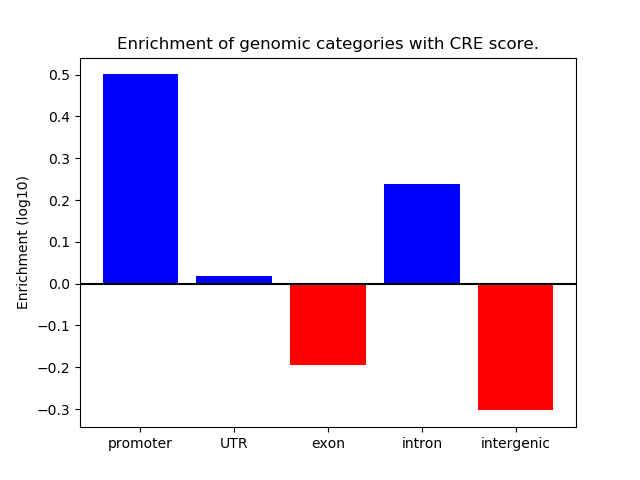
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr16_50309458_50309740 | Bbx | 21394 | 0.272779 | -0.47 | 3.1e-04 | Click! |
| chr16_50309820_50310005 | Bbx | 21081 | 0.273717 | -0.37 | 4.9e-03 | Click! |
| chr16_50381432_50381608 | Bbx | 48392 | 0.178932 | -0.37 | 5.1e-03 | Click! |
| chr16_50381060_50381246 | Bbx | 48759 | 0.177910 | -0.34 | 1.0e-02 | Click! |
| chr16_50221248_50221422 | Bbx | 109658 | 0.068622 | -0.28 | 3.6e-02 | Click! |
Activity of the Bbx motif across conditions
Conditions sorted by the z-value of the Bbx motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
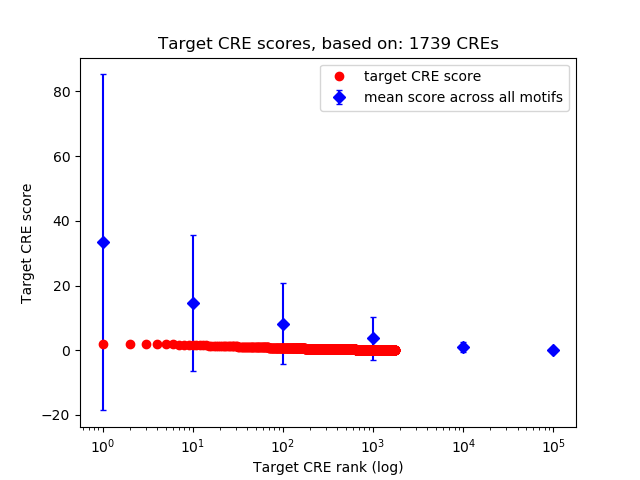
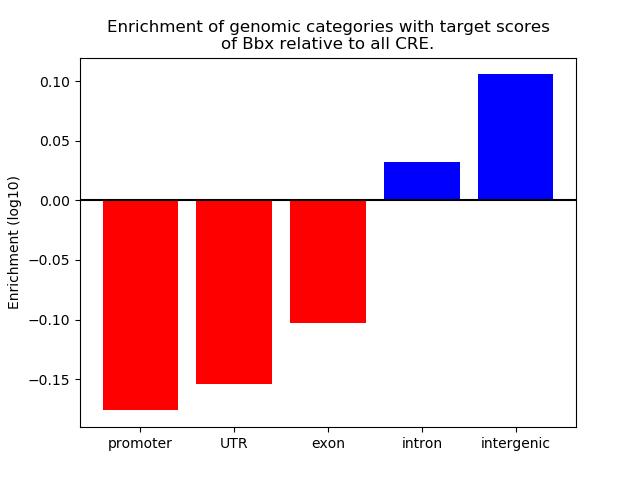
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_121348775_121349004 | 2.05 |
Hs3st2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
42970 |
0.13 |
| chrX_102418826_102418977 | 2.01 |
Hdac8 |
histone deacetylase 8 |
85840 |
0.07 |
| chr7_87371496_87371647 | 2.00 |
Tyr |
tyrosinase |
121821 |
0.05 |
| chr2_84338187_84338338 | 1.97 |
Gm13711 |
predicted gene 13711 |
19951 |
0.19 |
| chr4_49548338_49548489 | 1.93 |
Aldob |
aldolase B, fructose-bisphosphate |
1133 |
0.41 |
| chrX_9256889_9257156 | 1.93 |
Gm14862 |
predicted gene 14862 |
123 |
0.95 |
| chr13_21924698_21924984 | 1.68 |
Gm44456 |
predicted gene, 44456 |
175 |
0.85 |
| chr9_102656851_102657023 | 1.66 |
Anapc13 |
anaphase promoting complex subunit 13 |
28425 |
0.1 |
| chr19_11724358_11724660 | 1.66 |
Gm19224 |
predicted gene, 19224 |
20408 |
0.08 |
| chr17_57796735_57796897 | 1.65 |
Cntnap5c |
contactin associated protein-like 5C |
27246 |
0.14 |
| chr6_128551311_128551462 | 1.65 |
A2ml1 |
alpha-2-macroglobulin like 1 |
1892 |
0.19 |
| chr10_100155713_100155864 | 1.60 |
Gm25287 |
predicted gene, 25287 |
11534 |
0.13 |
| chr10_45001413_45001580 | 1.54 |
Gm4795 |
predicted pseudogene 4795 |
4614 |
0.19 |
| chr8_128424742_128424907 | 1.48 |
Nrp1 |
neuropilin 1 |
65427 |
0.12 |
| chr2_151037749_151037912 | 1.43 |
Nanp |
N-acetylneuraminic acid phosphatase |
1532 |
0.22 |
| chr1_163477892_163478043 | 1.42 |
Gorab |
golgin, RAB6-interacting |
74298 |
0.09 |
| chr2_171633456_171634007 | 1.39 |
Gm26489 |
predicted gene, 26489 |
327300 |
0.01 |
| chr2_151563537_151563699 | 1.39 |
Sdcbp2 |
syndecan binding protein (syntenin) 2 |
9004 |
0.13 |
| chr6_30477529_30477700 | 1.38 |
Tmem209 |
transmembrane protein 209 |
14565 |
0.12 |
| chr11_101556560_101556853 | 1.35 |
Nbr1 |
NBR1, autophagy cargo receptor |
534 |
0.6 |
| chr3_80780882_80781033 | 1.35 |
Gria2 |
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
21622 |
0.23 |
| chr5_14960858_14961009 | 1.29 |
Gm10354 |
predicted gene 10354 |
18002 |
0.18 |
| chr4_145000143_145000472 | 1.28 |
Vps13d |
vacuolar protein sorting 13D |
16346 |
0.21 |
| chr15_47761459_47761610 | 1.25 |
Gm23530 |
predicted gene, 23530 |
39139 |
0.19 |
| chr4_121014564_121014855 | 1.24 |
Smap2 |
small ArfGAP 2 |
2538 |
0.19 |
| chr2_167492358_167493413 | 1.21 |
Spata2 |
spermatogenesis associated 2 |
2 |
0.76 |
| chr13_91807162_91808007 | 1.19 |
Zcchc9 |
zinc finger, CCHC domain containing 9 |
77 |
0.97 |
| chr16_23511404_23511561 | 1.19 |
Gm49514 |
predicted gene, 49514 |
2788 |
0.2 |
| chr4_95141587_95141785 | 1.18 |
Gm28096 |
predicted gene 28096 |
10054 |
0.16 |
| chr17_63979339_63979498 | 1.15 |
Fer |
fer (fms/fps related) protein kinase |
41446 |
0.2 |
| chr10_36484838_36485024 | 1.15 |
Hs3st5 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 5 |
21883 |
0.26 |
| chr9_32005374_32005525 | 1.15 |
Gm47465 |
predicted gene, 47465 |
11 |
0.97 |
| chr6_135325338_135325489 | 1.14 |
Gm44275 |
predicted gene, 44275 |
5852 |
0.13 |
| chr6_103616019_103616201 | 1.14 |
Gm44436 |
predicted gene, 44436 |
147 |
0.95 |
| chr16_4100574_4100725 | 1.13 |
Crebbp |
CREB binding protein |
4523 |
0.15 |
| chr10_123089632_123089795 | 1.12 |
Mon2 |
MON2 homolog, regulator of endosome to Golgi trafficking |
13208 |
0.19 |
| chr6_139882188_139882339 | 1.09 |
Pik3c2g |
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
35720 |
0.16 |
| chr7_5060805_5061447 | 1.08 |
Gm45133 |
predicted gene 45133 |
923 |
0.22 |
| chr14_87566978_87567524 | 1.07 |
Tdrd3 |
tudor domain containing 3 |
28152 |
0.2 |
| chr10_61412962_61413255 | 1.06 |
Nodal |
nodal |
4864 |
0.13 |
| chr5_99099723_99099909 | 1.05 |
Prkg2 |
protein kinase, cGMP-dependent, type II |
62465 |
0.11 |
| chr9_110422694_110422845 | 1.05 |
Ngp |
neutrophilic granule protein |
3022 |
0.17 |
| chr10_99189347_99189498 | 1.05 |
Poc1b |
POC1 centriolar protein B |
3470 |
0.16 |
| chr2_174291748_174291899 | 1.04 |
Gnasas1 |
GNAS antisense RNA 1 |
3566 |
0.16 |
| chr17_64961925_64962076 | 1.04 |
Gm25344 |
predicted gene, 25344 |
17148 |
0.25 |
| chr3_60168628_60168801 | 1.04 |
Gm24382 |
predicted gene, 24382 |
40967 |
0.17 |
| chr1_175028715_175029238 | 1.03 |
Rgs7 |
regulator of G protein signaling 7 |
49969 |
0.17 |
| chr3_60169438_60169604 | 1.03 |
Gm24382 |
predicted gene, 24382 |
41774 |
0.16 |
| chr4_142221224_142221954 | 1.02 |
Kazn |
kazrin, periplakin interacting protein |
11383 |
0.22 |
| chr10_68444362_68444513 | 1.02 |
Cabcoco1 |
ciliary associated calcium binding coiled-coil 1 |
81330 |
0.09 |
| chr5_108451688_108451839 | 1.02 |
Mfsd7a |
major facilitator superfamily domain containing 7A |
2663 |
0.15 |
| chr4_138938899_138939064 | 1.00 |
Otud3 |
OTU domain containing 3 |
25036 |
0.12 |
| chr16_72510448_72511408 | 0.99 |
Robo1 |
roundabout guidance receptor 1 |
52772 |
0.18 |
| chr13_21170243_21170544 | 0.98 |
Trim27 |
tripartite motif-containing 27 |
9052 |
0.13 |
| chr1_191882872_191883023 | 0.98 |
1700034H15Rik |
RIKEN cDNA 1700034H15 gene |
16182 |
0.14 |
| chr17_6731741_6732102 | 0.97 |
Ezr |
ezrin |
10900 |
0.16 |
| chr3_83015499_83015650 | 0.97 |
Gm30097 |
predicted gene, 30097 |
7086 |
0.14 |
| chr12_46698300_46698451 | 0.95 |
Gm33869 |
predicted gene, 33869 |
5841 |
0.15 |
| chr13_75385061_75385212 | 0.92 |
Gm48234 |
predicted gene, 48234 |
94251 |
0.07 |
| chr12_54256573_54256724 | 0.91 |
Gm24692 |
predicted gene, 24692 |
15748 |
0.14 |
| chr6_90561220_90561762 | 0.91 |
Aldh1l1 |
aldehyde dehydrogenase 1 family, member L1 |
6215 |
0.15 |
| chr13_100409933_100410098 | 0.90 |
Rpl31-ps13 |
ribosomal protein L31, pseudogene 13 |
5550 |
0.15 |
| chr9_61370339_61371660 | 0.89 |
Gm10655 |
predicted gene 10655 |
628 |
0.63 |
| chr8_46446597_46446889 | 0.88 |
Gm45245 |
predicted gene 45245 |
5561 |
0.16 |
| chr3_144201827_144203140 | 0.88 |
Lmo4 |
LIM domain only 4 |
87 |
0.97 |
| chr13_7698051_7698203 | 0.88 |
Gm48252 |
predicted gene, 48252 |
55151 |
0.14 |
| chr14_14351950_14353283 | 0.87 |
Il3ra |
interleukin 3 receptor, alpha chain |
2995 |
0.15 |
| chr12_100725043_100725700 | 0.86 |
Rps6ka5 |
ribosomal protein S6 kinase, polypeptide 5 |
251 |
0.91 |
| chr2_137880540_137880691 | 0.85 |
Gm14062 |
predicted gene 14062 |
3784 |
0.38 |
| chrX_16836560_16836772 | 0.83 |
Maob |
monoamine oxidase B |
19300 |
0.26 |
| chr6_82052849_82053004 | 0.83 |
Gm15864 |
predicted gene 15864 |
345 |
0.67 |
| chr5_91038675_91038962 | 0.83 |
Epgn |
epithelial mitogen |
11354 |
0.17 |
| chr7_111003851_111004038 | 0.82 |
Mrvi1 |
MRV integration site 1 |
21483 |
0.15 |
| chr14_99305409_99305608 | 0.82 |
Klf5 |
Kruppel-like factor 5 |
6354 |
0.17 |
| chr18_84857828_84857986 | 0.82 |
Gm16146 |
predicted gene 16146 |
1640 |
0.31 |
| chr5_15019515_15019782 | 0.82 |
Gm17019 |
predicted gene 17019 |
13350 |
0.22 |
| chr11_44683091_44683242 | 0.82 |
Gm12158 |
predicted gene 12158 |
20490 |
0.2 |
| chr14_76805109_76805269 | 0.82 |
Gm30246 |
predicted gene, 30246 |
24362 |
0.16 |
| chr16_32563013_32563416 | 0.81 |
Gm34680 |
predicted gene, 34680 |
11472 |
0.14 |
| chr11_106274798_106274991 | 0.80 |
Tcam1 |
testicular cell adhesion molecule 1 |
1778 |
0.17 |
| chr13_100284737_100284888 | 0.79 |
Naip3-ps1 |
NLR family, apoptosis inhibitory protein 3, pseudogene 1 |
5995 |
0.17 |
| chr3_67867676_67867912 | 0.79 |
Iqcj |
IQ motif containing J |
24426 |
0.19 |
| chr1_85333250_85333459 | 0.78 |
Gm2619 |
predicted gene 2619 |
6069 |
0.14 |
| chr2_6285591_6285742 | 0.78 |
Gm13383 |
predicted gene 13383 |
28400 |
0.14 |
| chr2_148393301_148393467 | 0.78 |
Sstr4 |
somatostatin receptor 4 |
1960 |
0.3 |
| chr12_80112847_80113228 | 0.77 |
Zfp36l1 |
zinc finger protein 36, C3H type-like 1 |
24 |
0.96 |
| chr5_15591695_15592053 | 0.77 |
Gm30613 |
predicted gene, 30613 |
13192 |
0.14 |
| chr5_137472693_137472990 | 0.77 |
Zan |
zonadhesin |
3754 |
0.11 |
| chr16_26991829_26992040 | 0.76 |
Gmnc |
geminin coiled-coil domain containing |
282 |
0.95 |
| chr13_19364615_19364766 | 0.76 |
Stard3nl |
STARD3 N-terminal like |
3265 |
0.26 |
| chr17_3556298_3556461 | 0.76 |
Tfb1m |
transcription factor B1, mitochondrial |
1355 |
0.33 |
| chr13_56139567_56139718 | 0.76 |
Macroh2a1 |
macroH2A.1 histone |
3281 |
0.22 |
| chr9_124425359_124425530 | 0.75 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
1290 |
0.39 |
| chr7_99582614_99582765 | 0.75 |
Arrb1 |
arrestin, beta 1 |
9625 |
0.11 |
| chr12_111353338_111354089 | 0.74 |
Cdc42bpb |
CDC42 binding protein kinase beta |
23906 |
0.13 |
| chr8_76913114_76913300 | 0.73 |
Nr3c2 |
nuclear receptor subfamily 3, group C, member 2 |
4935 |
0.24 |
| chr7_55332148_55332299 | 0.73 |
Gm44898 |
predicted gene 44898 |
56287 |
0.13 |
| chr3_113699092_113699243 | 0.72 |
Gm17775 |
predicted gene, 17775 |
30065 |
0.19 |
| chr14_14354416_14355184 | 0.72 |
Il3ra |
interleukin 3 receptor, alpha chain |
5179 |
0.12 |
| chr6_67013417_67013625 | 0.72 |
Gm15644 |
predicted gene 15644 |
1036 |
0.38 |
| chr19_4593871_4594048 | 0.71 |
Pcx |
pyruvate carboxylase |
387 |
0.78 |
| chr7_128003368_128003519 | 0.71 |
Trim72 |
tripartite motif-containing 72 |
506 |
0.52 |
| chr18_25097355_25097753 | 0.71 |
Fhod3 |
formin homology 2 domain containing 3 |
26807 |
0.22 |
| chr5_147758193_147758351 | 0.71 |
Gm43156 |
predicted gene 43156 |
12756 |
0.19 |
| chr13_90502235_90502410 | 0.70 |
Gm21726 |
predicted gene, 21726 |
81007 |
0.09 |
| chr4_19970756_19970907 | 0.70 |
4930480G23Rik |
RIKEN cDNA 4930480G23 gene |
27893 |
0.17 |
| chr13_114793412_114793586 | 0.70 |
Mocs2 |
molybdenum cofactor synthesis 2 |
24737 |
0.23 |
| chr4_77310465_77310616 | 0.69 |
Gm23159 |
predicted gene, 23159 |
352042 |
0.01 |
| chr3_155777141_155777292 | 0.69 |
Gm37324 |
predicted gene, 37324 |
1388 |
0.43 |
| chr15_59520085_59520242 | 0.69 |
Gm36677 |
predicted gene, 36677 |
98427 |
0.07 |
| chr16_52333248_52333399 | 0.69 |
Alcam |
activated leukocyte cell adhesion molecule |
36399 |
0.23 |
| chr2_24720034_24720185 | 0.69 |
Cacna1b |
calcium channel, voltage-dependent, N type, alpha 1B subunit |
34241 |
0.13 |
| chr5_134914340_134914552 | 0.67 |
Cldn13 |
claudin 13 |
1080 |
0.29 |
| chr1_168295882_168296042 | 0.67 |
Gm37524 |
predicted gene, 37524 |
41709 |
0.18 |
| chr5_50794878_50795077 | 0.67 |
Gm43014 |
predicted gene 43014 |
43340 |
0.19 |
| chr18_25655666_25655955 | 0.67 |
0710001A04Rik |
RIKEN cDNA 0710001A04 gene |
57960 |
0.13 |
| chr4_99249480_99249645 | 0.66 |
Gm10305 |
predicted gene 10305 |
23109 |
0.15 |
| chr15_101177114_101177265 | 0.66 |
Acvr1b |
activin A receptor, type 1B |
3122 |
0.15 |
| chr19_47053280_47053431 | 0.66 |
Gm50319 |
predicted gene, 50319 |
1893 |
0.18 |
| chr12_80114135_80114615 | 0.66 |
Zfp36l1 |
zinc finger protein 36, C3H type-like 1 |
1362 |
0.27 |
| chr11_56069627_56069962 | 0.65 |
Nmur2 |
neuromedin U receptor 2 |
28784 |
0.25 |
| chr5_146021259_146021424 | 0.65 |
Gm8454 |
predicted gene 8454 |
3500 |
0.16 |
| chr9_71164075_71164226 | 0.64 |
Gm32511 |
predicted gene, 32511 |
356 |
0.75 |
| chr4_89433459_89434002 | 0.64 |
Gm12608 |
predicted gene 12608 |
10914 |
0.19 |
| chr15_63709612_63709777 | 0.63 |
4930449C09Rik |
RIKEN cDNA 4930449C09 gene |
8124 |
0.14 |
| chr8_68613276_68613575 | 0.63 |
Gm15654 |
predicted gene 15654 |
6713 |
0.28 |
| chr7_123448677_123449086 | 0.63 |
Aqp8 |
aquaporin 8 |
13410 |
0.18 |
| chr3_106495488_106495639 | 0.63 |
Dennd2d |
DENN/MADD domain containing 2D |
2388 |
0.21 |
| chr10_111073345_111073645 | 0.63 |
Gm48851 |
predicted gene, 48851 |
29376 |
0.14 |
| chr12_111246912_111247092 | 0.62 |
Traf3 |
TNF receptor-associated factor 3 |
1481 |
0.35 |
| chr8_127514893_127515068 | 0.62 |
Gm6921 |
predicted pseudogene 6921 |
46455 |
0.16 |
| chr5_62622956_62623107 | 0.61 |
Gm43237 |
predicted gene 43237 |
9573 |
0.2 |
| chr8_8225399_8225550 | 0.61 |
A630009H07Rik |
RIKEN cDNA A630009H07 gene |
109603 |
0.06 |
| chr11_22055931_22056082 | 0.61 |
Otx1 |
orthodenticle homeobox 1 |
53109 |
0.15 |
| chr13_63567343_63568686 | 0.60 |
A930032L01Rik |
RIKEN cDNA A930032L01 gene |
40 |
0.96 |
| chr10_62530548_62530699 | 0.60 |
Srgn |
serglycin |
3172 |
0.18 |
| chr4_133552955_133553552 | 0.60 |
Nr0b2 |
nuclear receptor subfamily 0, group B, member 2 |
123 |
0.92 |
| chr13_95531340_95531619 | 0.60 |
Gm48730 |
predicted gene, 48730 |
4111 |
0.18 |
| chr10_44427597_44427753 | 0.60 |
Gm35028 |
predicted gene, 35028 |
3351 |
0.25 |
| chr5_41842836_41842987 | 0.59 |
Bod1l |
biorientation of chromosomes in cell division 1-like |
1381 |
0.44 |
| chr11_112720530_112720814 | 0.59 |
BC006965 |
cDNA sequence BC006965 |
9309 |
0.26 |
| chr19_10277406_10277748 | 0.59 |
Gm30042 |
predicted gene, 30042 |
13349 |
0.13 |
| chr10_121604767_121604918 | 0.59 |
Xpot |
exportin, tRNA (nuclear export receptor for tRNAs) |
244 |
0.9 |
| chr3_14877046_14877264 | 0.58 |
Car3 |
carbonic anhydrase 3 |
7977 |
0.19 |
| chr12_57537131_57537289 | 0.58 |
Foxa1 |
forkhead box A1 |
8911 |
0.15 |
| chr1_91442484_91442930 | 0.58 |
Per2 |
period circadian clock 2 |
7158 |
0.12 |
| chr4_131041413_131041564 | 0.58 |
A930031H19Rik |
RIKEN cDNA A930031H19 gene |
31014 |
0.18 |
| chr1_139096497_139096648 | 0.58 |
Gm37655 |
predicted gene, 37655 |
9588 |
0.13 |
| chr4_7567873_7568075 | 0.57 |
8430436N08Rik |
RIKEN cDNA 8430436N08 gene |
7286 |
0.32 |
| chr4_141131586_141132124 | 0.57 |
Szrd1 |
SUZ RNA binding domain containing 1 |
7872 |
0.11 |
| chr2_133511465_133511774 | 0.57 |
Bmp2 |
bone morphogenetic protein 2 |
40540 |
0.14 |
| chr16_44133385_44133536 | 0.57 |
Atp6v1a |
ATPase, H+ transporting, lysosomal V1 subunit A |
5520 |
0.19 |
| chr4_11762874_11763219 | 0.57 |
Cdh17 |
cadherin 17 |
4853 |
0.25 |
| chr17_36894286_36894452 | 0.57 |
Trim31 |
tripartite motif-containing 31 |
3749 |
0.1 |
| chr1_13660790_13660970 | 0.56 |
Lactb2 |
lactamase, beta 2 |
334 |
0.87 |
| chr11_33203073_33203919 | 0.56 |
Tlx3 |
T cell leukemia, homeobox 3 |
93 |
0.88 |
| chr8_22025214_22025365 | 0.56 |
Atp7b |
ATPase, Cu++ transporting, beta polypeptide |
34730 |
0.09 |
| chr4_97102030_97102181 | 0.56 |
Gm27521 |
predicted gene, 27521 |
185085 |
0.03 |
| chr12_12042917_12043068 | 0.56 |
Gm4294 |
predicted gene 4294 |
19947 |
0.2 |
| chr2_153218461_153218795 | 0.56 |
Tspyl3 |
TSPY-like 3 |
6813 |
0.14 |
| chr8_80055516_80055926 | 0.56 |
Hhip |
Hedgehog-interacting protein |
2285 |
0.32 |
| chr3_52254073_52254404 | 0.56 |
Olfr1398-ps1 |
olfactory receptor 1398, pseudogene 1 |
7293 |
0.13 |
| chr4_135874474_135874625 | 0.56 |
Pnrc2 |
proline-rich nuclear receptor coactivator 2 |
699 |
0.52 |
| chr3_80787470_80787641 | 0.56 |
Gria2 |
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
15024 |
0.25 |
| chr2_129241136_129241588 | 0.55 |
Gm14024 |
predicted gene 14024 |
5014 |
0.11 |
| chr7_72826599_72826750 | 0.55 |
Gm7693 |
predicted gene 7693 |
113053 |
0.06 |
| chr1_67186231_67186382 | 0.55 |
Gm15668 |
predicted gene 15668 |
62894 |
0.11 |
| chr7_126630054_126630205 | 0.55 |
Nupr1 |
nuclear protein transcription regulator 1 |
732 |
0.42 |
| chr3_29732889_29733040 | 0.54 |
Gm37557 |
predicted gene, 37557 |
38320 |
0.19 |
| chr13_62888125_62888325 | 0.54 |
Fbp1 |
fructose bisphosphatase 1 |
57 |
0.97 |
| chr18_62211148_62211299 | 0.54 |
Gm9949 |
predicted gene 9949 |
31097 |
0.17 |
| chr10_68090256_68090550 | 0.54 |
Arid5b |
AT rich interactive domain 5B (MRF1-like) |
46223 |
0.13 |
| chrX_9222912_9223090 | 0.54 |
Lancl3 |
LanC lantibiotic synthetase component C-like 3 (bacterial) |
23099 |
0.12 |
| chr4_149445330_149446172 | 0.54 |
Rbp7 |
retinol binding protein 7, cellular |
7673 |
0.11 |
| chr11_98968695_98969422 | 0.53 |
Rara |
retinoic acid receptor, alpha |
8646 |
0.1 |
| chr6_59162029_59162180 | 0.53 |
Gm43905 |
predicted gene, 43905 |
31149 |
0.17 |
| chr8_3252706_3253021 | 0.53 |
Gm16180 |
predicted gene 16180 |
6841 |
0.21 |
| chr3_20027390_20027663 | 0.53 |
Hps3 |
HPS3, biogenesis of lysosomal organelles complex 2 subunit 1 |
7754 |
0.19 |
| chr15_78413114_78413438 | 0.52 |
Mpst |
mercaptopyruvate sulfurtransferase |
3294 |
0.12 |
| chr7_135581161_135581493 | 0.52 |
Ptpre |
protein tyrosine phosphatase, receptor type, E |
12061 |
0.17 |
| chr17_94094392_94094543 | 0.52 |
Gm50004 |
predicted gene, 50004 |
37804 |
0.23 |
| chr16_44177510_44177675 | 0.52 |
Usf3 |
upstream transcription factor family member 3 |
4195 |
0.23 |
| chr19_10166053_10166204 | 0.52 |
Gm50359 |
predicted gene, 50359 |
10033 |
0.11 |
| chr7_128289110_128289522 | 0.51 |
BC017158 |
cDNA sequence BC017158 |
611 |
0.44 |
| chr1_131310511_131310734 | 0.51 |
Srgap2 |
SLIT-ROBO Rho GTPase activating protein 2 |
443 |
0.76 |
| chrX_166627284_166627435 | 0.51 |
Gm15226 |
predicted gene 15226 |
5817 |
0.21 |
| chr1_90481315_90481666 | 0.51 |
Gm28722 |
predicted gene 28722 |
26989 |
0.19 |
| chr14_121214755_121214906 | 0.51 |
B930095G15Rik |
RIKEN cDNA B930095G15 gene |
33876 |
0.17 |
| chr4_57954719_57954870 | 0.51 |
Txn1 |
thioredoxin 1 |
1617 |
0.4 |
| chr5_97007576_97007868 | 0.51 |
Bmp2k |
BMP2 inducible kinase |
10033 |
0.14 |
| chr7_84040262_84040413 | 0.51 |
Cemip |
cell migration inducing protein, hyaluronan binding |
46165 |
0.13 |
| chr1_13655707_13655872 | 0.50 |
Lactb2 |
lactamase, beta 2 |
4757 |
0.19 |
| chr13_31531222_31531375 | 0.50 |
Foxq1 |
forkhead box Q1 |
24836 |
0.13 |
| chr19_11022451_11022797 | 0.50 |
Ms4a18 |
membrane-spanning 4-domains, subfamily A, member 18 |
4593 |
0.12 |
| chr4_69594168_69594334 | 0.50 |
Gm11221 |
predicted gene 11221 |
97727 |
0.09 |
| chr4_43964689_43964890 | 0.50 |
Glipr2 |
GLI pathogenesis-related 2 |
7097 |
0.14 |
| chr18_13943186_13943337 | 0.50 |
Zfp521 |
zinc finger protein 521 |
28459 |
0.21 |
| chr6_17554824_17555153 | 0.49 |
Met |
met proto-oncogene |
8015 |
0.24 |
| chr18_19524329_19524663 | 0.49 |
Gm7720 |
predicted gene 7720 |
25897 |
0.27 |
| chr5_145621725_145621876 | 0.49 |
Selenok-ps7 |
selenoprotein K, pseudogene 7 |
1567 |
0.33 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.3 | 0.8 | GO:0061625 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.1 | 0.4 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.5 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.1 | 0.9 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 0.5 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.6 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.3 | GO:0050904 | diapedesis(GO:0050904) |
| 0.1 | 0.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.4 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.1 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 1.0 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.1 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.2 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.2 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.1 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.3 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.2 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 0.2 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 0.2 | GO:0043309 | regulation of eosinophil degranulation(GO:0043309) |
| 0.1 | 0.2 | GO:0032513 | negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 | 0.1 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.1 | 0.3 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 | 0.2 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.5 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.4 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:0006548 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 1.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.4 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.2 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.2 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.2 | GO:0006971 | hypotonic response(GO:0006971) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.4 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:1901563 | response to camptothecin(GO:1901563) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.3 | GO:0031269 | pseudopodium assembly(GO:0031269) |
| 0.0 | 0.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.6 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.0 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.0 | 0.3 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0070668 | mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.0 | 0.2 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.0 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.0 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.1 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.2 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:0060336 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0044004 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.1 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.3 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 0.0 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 0.1 | GO:0071455 | cellular response to increased oxygen levels(GO:0036295) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.0 | GO:0009757 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.1 | GO:0090241 | negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.0 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.0 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.0 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.0 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 0.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.4 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.0 | GO:1904193 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.0 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.0 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.0 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 1.0 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.0 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.0 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:0009115 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.0 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 0.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.0 | GO:0042505 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.0 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.0 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.3 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.1 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.0 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.0 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.0 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.0 | GO:0072093 | metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 0.0 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.0 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.1 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.1 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.3 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.1 | GO:0034377 | plasma lipoprotein particle assembly(GO:0034377) |
| 0.0 | 0.1 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.0 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0002923 | regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002923) |
| 0.0 | 0.1 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.3 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.0 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.0 | 0.2 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.0 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.0 | GO:2000561 | regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0097006 | regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.0 | 0.0 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.0 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.1 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:1902224 | ketone body metabolic process(GO:1902224) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.0 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.0 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.0 | 0.0 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.0 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.0 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.0 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.1 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.0 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.0 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.0 | 0.0 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.5 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.1 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.0 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.0 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 1.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 0.2 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.4 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.2 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.1 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.2 | GO:0052796 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.0 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0019145 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0051380 | beta-adrenergic receptor activity(GO:0004939) norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.6 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.0 | GO:0030792 | methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.0 | GO:0030351 | inositol-1,3,4,5,6-pentakisphosphate 3-phosphatase activity(GO:0030351) inositol-1,4,5,6-tetrakisphosphate 6-phosphatase activity(GO:0030352) inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.0 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.0 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.0 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.0 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.8 | GO:0018423 | protein C-terminal leucine carboxyl O-methyltransferase activity(GO:0018423) |
| 0.0 | 0.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0015254 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.0 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.0 | 0.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 0.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.6 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.0 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |