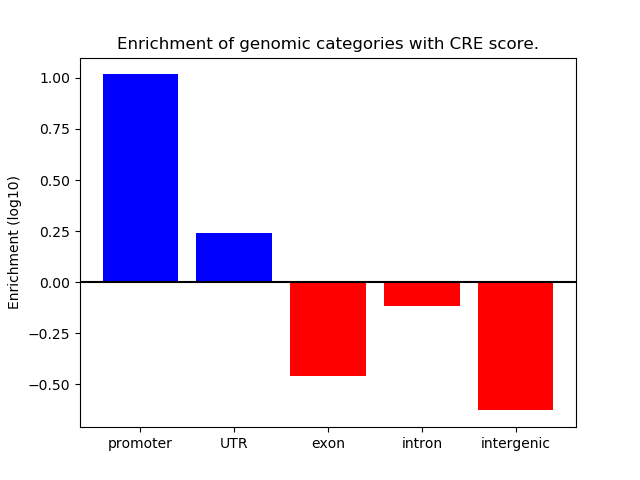
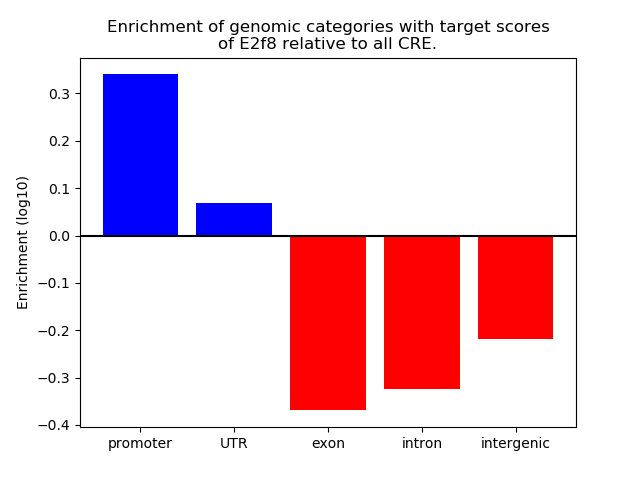
Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
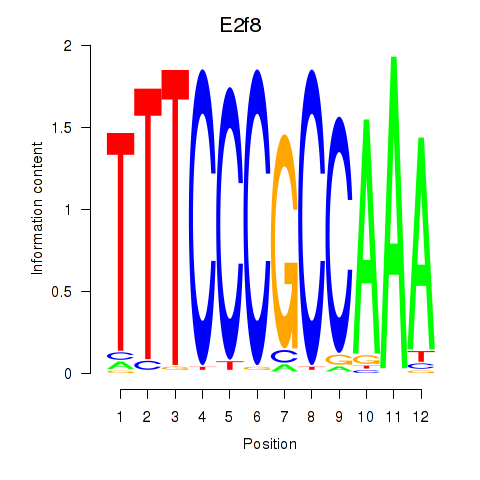
Results for E2f8
Z-value: 0.73
Transcription factors associated with E2f8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f8
|
ENSMUSG00000046179.11 | E2F transcription factor 8 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_48882387_48882677 | E2f8 | 936 | 0.381853 | 0.78 | 3.7e-12 | Click! |
| chr7_48879698_48879867 | E2f8 | 1222 | 0.313240 | 0.68 | 1.0e-08 | Click! |
| chr7_48880593_48880868 | E2f8 | 274 | 0.704743 | 0.66 | 5.0e-08 | Click! |
| chr7_48882737_48882888 | E2f8 | 1216 | 0.313490 | 0.49 | 1.6e-04 | Click! |
| chr7_48875128_48875279 | E2f8 | 48 | 0.961423 | -0.02 | 8.8e-01 | Click! |
Activity of the E2f8 motif across conditions
Conditions sorted by the z-value of the E2f8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
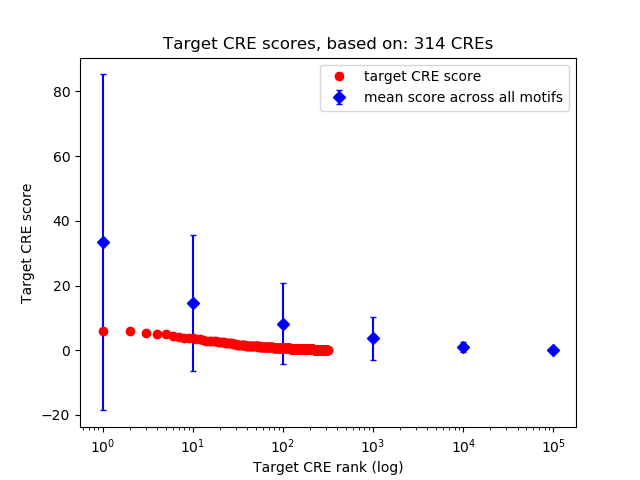
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_48545202_48545700 | 5.86 |
Sorcs3 |
sortilin-related VPS10 domain containing receptor 3 |
115491 |
0.07 |
| chr9_100643446_100643943 | 5.86 |
Stag1 |
stromal antigen 1 |
58 |
0.97 |
| chr15_55089779_55091008 | 5.32 |
Dscc1 |
DNA replication and sister chromatid cohesion 1 |
87 |
0.96 |
| chr9_115165935_115166094 | 5.10 |
Gm4665 |
predicted gene 4665 |
20044 |
0.15 |
| chr3_123432402_123432553 | 4.98 |
Prss12 |
protease, serine 12 neurotrypsin (motopsin) |
14436 |
0.13 |
| chr5_150673477_150673662 | 4.34 |
Pds5b |
PDS5 cohesin associated factor B |
170 |
0.92 |
| chr12_24708357_24708959 | 3.93 |
Rrm2 |
ribonucleotide reductase M2 |
94 |
0.96 |
| chr9_36725848_36726788 | 3.79 |
Chek1 |
checkpoint kinase 1 |
56 |
0.96 |
| chr3_102145742_102145917 | 3.65 |
Casq2 |
calsequestrin 2 |
1778 |
0.26 |
| chr12_104404094_104404246 | 3.64 |
Serpina3n |
serine (or cysteine) peptidase inhibitor, clade A, member 3N |
2559 |
0.19 |
| chr3_95665078_95665320 | 3.50 |
Mcl1 |
myeloid cell leukemia sequence 1 |
6342 |
0.11 |
| chr8_23034325_23034515 | 3.49 |
Ank1 |
ankyrin 1, erythroid |
679 |
0.69 |
| chr2_25195736_25196168 | 3.13 |
Tor4a |
torsin family 4, member A |
807 |
0.31 |
| chr16_10834942_10835141 | 2.86 |
Rmi2 |
RecQ mediated genome instability 2 |
18 |
0.95 |
| chr17_56303166_56303753 | 2.84 |
Uhrf1 |
ubiquitin-like, containing PHD and RING finger domains, 1 |
59 |
0.95 |
| chr4_137795768_137796555 | 2.77 |
Alpl |
alkaline phosphatase, liver/bone/kidney |
69 |
0.98 |
| chr11_98907626_98907840 | 2.74 |
Cdc6 |
cell division cycle 6 |
68 |
0.95 |
| chr6_34780285_34780491 | 2.71 |
Agbl3 |
ATP/GTP binding protein-like 3 |
44 |
0.96 |
| chr7_121981252_121981861 | 2.63 |
Cog7 |
component of oligomeric golgi complex 7 |
138 |
0.59 |
| chr15_61985341_61985712 | 2.50 |
Myc |
myelocytomatosis oncogene |
38 |
0.98 |
| chr8_45253145_45253404 | 2.44 |
F11 |
coagulation factor XI |
8757 |
0.18 |
| chr4_72182335_72182783 | 2.41 |
Gm11250 |
predicted gene 11250 |
13791 |
0.19 |
| chr13_73475366_73476070 | 2.35 |
Lpcat1 |
lysophosphatidylcholine acyltransferase 1 |
3967 |
0.26 |
| chr9_51008308_51009431 | 2.29 |
Sik2 |
salt inducible kinase 2 |
67 |
0.97 |
| chr12_24706721_24707100 | 2.26 |
Rrm2 |
ribonucleotide reductase M2 |
1331 |
0.35 |
| chr5_137684632_137685345 | 2.12 |
Agfg2 |
ArfGAP with FG repeats 2 |
262 |
0.81 |
| chr6_31586330_31586508 | 2.08 |
Gm6117 |
predicted gene 6117 |
11452 |
0.17 |
| chr1_86581833_86582394 | 1.81 |
Pde6d |
phosphodiesterase 6D, cGMP-specific, rod, delta |
404 |
0.62 |
| chr2_75632701_75632852 | 1.79 |
Gm13655 |
predicted gene 13655 |
606 |
0.69 |
| chr11_29172269_29172975 | 1.79 |
Ppp4r3b |
protein phosphatase 4 regulatory subunit 3B |
268 |
0.83 |
| chr17_8272658_8272950 | 1.68 |
Mpc1 |
mitochondrial pyruvate carrier 1 |
10100 |
0.13 |
| chr8_109737190_109738048 | 1.67 |
Atxn1l |
ataxin 1-like |
103 |
0.96 |
| chr8_112010957_112011159 | 1.67 |
Kars |
lysyl-tRNA synthetase |
208 |
0.64 |
| chr12_54999125_54999276 | 1.61 |
Baz1a |
bromodomain adjacent to zinc finger domain 1A |
98 |
0.94 |
| chr11_26387078_26387273 | 1.59 |
Fancl |
Fanconi anemia, complementation group L |
27 |
0.98 |
| chr4_115062632_115062827 | 1.55 |
Tal1 |
T cell acute lymphocytic leukemia 1 |
3221 |
0.2 |
| chr2_130663705_130664395 | 1.49 |
Ddrgk1 |
DDRGK domain containing 1 |
115 |
0.93 |
| chr17_29398006_29398391 | 1.47 |
Fgd2 |
FYVE, RhoGEF and PH domain containing 2 |
21697 |
0.1 |
| chr5_23787844_23788302 | 1.45 |
Rint1 |
RAD50 interactor 1 |
332 |
0.77 |
| chr2_155374011_155374162 | 1.44 |
Trp53inp2 |
transformation related protein 53 inducible nuclear protein 2 |
6973 |
0.14 |
| chr13_101545392_101545560 | 1.44 |
Gm47533 |
predicted gene, 47533 |
270 |
0.89 |
| chr1_13573759_13574139 | 1.41 |
Tram1 |
translocating chain-associating membrane protein 1 |
5849 |
0.23 |
| chr11_16817466_16817617 | 1.41 |
Egfros |
epidermal growth factor receptor, opposite strand |
13161 |
0.21 |
| chr2_92371871_92372314 | 1.40 |
Large2 |
LARGE xylosyl- and glucuronyltransferase 2 |
125 |
0.92 |
| chrX_103622717_103623114 | 1.39 |
Ftx |
Ftx transcript, Xist regulator (non-protein coding) |
415 |
0.7 |
| chrX_105069770_105069963 | 1.37 |
5530601H04Rik |
RIKEN cDNA 5530601H04 gene |
169 |
0.94 |
| chr12_4233669_4233893 | 1.31 |
Cenpo |
centromere protein O |
157 |
0.6 |
| chr16_11270757_11270983 | 1.30 |
Mir1945 |
microRNA 1945 |
16425 |
0.12 |
| chr11_45805877_45806040 | 1.20 |
F630206G17Rik |
RIKEN cDNA F630206G17 gene |
2129 |
0.25 |
| chr2_60796909_60797719 | 1.20 |
Rbms1 |
RNA binding motif, single stranded interacting protein 1 |
16373 |
0.25 |
| chr14_20388519_20389160 | 1.16 |
Dnajc9 |
DnaJ heat shock protein family (Hsp40) member C9 |
18 |
0.95 |
| chr15_82340761_82340945 | 1.16 |
Pheta2 |
PH domain containing endocytic trafficking adaptor 2 |
5 |
0.61 |
| chr18_32556691_32557012 | 1.15 |
Gypc |
glycophorin C |
3129 |
0.27 |
| chr5_52189815_52190375 | 1.15 |
Dhx15 |
DEAH (Asp-Glu-Ala-His) box polypeptide 15 |
282 |
0.69 |
| chr1_72288975_72289126 | 1.14 |
Tmem169 |
transmembrane protein 169 |
4655 |
0.14 |
| chr13_81643178_81643470 | 1.14 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
10170 |
0.18 |
| chr17_71598263_71598670 | 1.14 |
Trmt61b |
tRNA methyltransferase 61B |
213 |
0.87 |
| chr5_75450662_75450882 | 1.09 |
Gm42800 |
predicted gene 42800 |
51400 |
0.13 |
| chr3_144198270_144199266 | 1.07 |
Gm43445 |
predicted gene 43445 |
494 |
0.79 |
| chr6_149310741_149310934 | 1.07 |
Resf1 |
retroelement silencing factor 1 |
579 |
0.7 |
| chr15_79690079_79691459 | 1.05 |
Gtpbp1 |
GTP binding protein 1 |
76 |
0.92 |
| chr11_88047219_88047407 | 1.05 |
Srsf1 |
serine and arginine-rich splicing factor 1 |
60 |
0.91 |
| chr12_53837518_53837703 | 0.99 |
1700060O08Rik |
RIKEN cDNA 1700060O08 gene |
241782 |
0.02 |
| chr12_110278889_110279209 | 0.99 |
Dio3 |
deiodinase, iodothyronine type III |
19 |
0.91 |
| chr2_103707189_103707351 | 0.98 |
Abtb2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
7765 |
0.17 |
| chr13_21783782_21783987 | 0.96 |
H3c11 |
H3 clustered histone 11 |
514 |
0.42 |
| chr6_100288289_100288440 | 0.93 |
Rybp |
RING1 and YY1 binding protein |
879 |
0.58 |
| chr19_38930513_38931703 | 0.93 |
Hells |
helicase, lymphoid specific |
66 |
0.98 |
| chr15_98092821_98093001 | 0.91 |
Senp1 |
SUMO1/sentrin specific peptidase 1 |
24 |
0.7 |
| chr11_54860143_54861186 | 0.90 |
Lyrm7 |
LYR motif containing 7 |
55 |
0.96 |
| chr16_16146270_16147094 | 0.90 |
Spidr |
scaffolding protein involved in DNA repair |
122 |
0.97 |
| chr16_32277357_32277552 | 0.89 |
Rnf168 |
ring finger protein 168 |
5 |
0.96 |
| chr17_17393048_17393199 | 0.88 |
Lix1 |
limb and CNS expressed 1 |
9549 |
0.14 |
| chr9_108569718_108570862 | 0.87 |
Dalrd3 |
DALR anticodon binding domain containing 3 |
376 |
0.53 |
| chr5_137293156_137293696 | 0.87 |
Mir8116 |
microRNA 8116 |
354 |
0.64 |
| chr11_102318737_102318932 | 0.87 |
Ubtf |
upstream binding transcription factor, RNA polymerase I |
262 |
0.85 |
| chr16_18248696_18249076 | 0.85 |
Trmt2a |
TRM2 tRNA methyltransferase 2A |
3 |
0.64 |
| chr7_123370222_123370701 | 0.83 |
Arhgap17 |
Rho GTPase activating protein 17 |
546 |
0.51 |
| chr12_119390165_119390727 | 0.83 |
Macc1 |
metastasis associated in colon cancer 1 |
71 |
0.98 |
| chr7_120865910_120866142 | 0.83 |
Gm15774 |
predicted gene 15774 |
9272 |
0.13 |
| chr2_128179990_128180452 | 0.82 |
Gm14009 |
predicted gene 14009 |
9766 |
0.22 |
| chr11_113785381_113786461 | 0.80 |
Sdk2 |
sidekick cell adhesion molecule 2 |
24502 |
0.14 |
| chr11_79992495_79993374 | 0.78 |
Suz12 |
SUZ12 polycomb repressive complex 2 subunit |
172 |
0.95 |
| chr11_120949321_120949700 | 0.78 |
Slc16a3 |
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
429 |
0.72 |
| chrX_150594244_150594395 | 0.78 |
Pfkfb1 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
159 |
0.94 |
| chrX_73969838_73969995 | 0.78 |
Hcfc1 |
host cell factor C1 |
3559 |
0.14 |
| chr9_35116070_35117303 | 0.78 |
4930581F22Rik |
RIKEN cDNA 4930581F22 gene |
42 |
0.69 |
| chr1_156986305_156986503 | 0.76 |
4930439D14Rik |
RIKEN cDNA 4930439D14 gene |
46566 |
0.11 |
| chr7_141658277_141658620 | 0.75 |
Muc6 |
mucin 6, gastric |
3129 |
0.19 |
| chr2_84743716_84744451 | 0.75 |
Gm19426 |
predicted gene, 19426 |
428 |
0.65 |
| chr16_55893491_55893669 | 0.75 |
Nxpe3 |
neurexophilin and PC-esterase domain family, member 3 |
1705 |
0.27 |
| chr6_145124644_145124840 | 0.75 |
Lrmp |
lymphoid-restricted membrane protein |
2961 |
0.19 |
| chr15_103014087_103015222 | 0.75 |
Mir615 |
microRNA 615 |
256 |
0.74 |
| chr14_43278790_43278941 | 0.74 |
Gm17153 |
predicted gene 17153 |
780 |
0.43 |
| chr12_12941527_12942979 | 0.74 |
Mycn |
v-myc avian myelocytomatosis viral related oncogene, neuroblastoma derived |
339 |
0.83 |
| chr6_142780696_142780976 | 0.72 |
Cmas |
cytidine monophospho-N-acetylneuraminic acid synthetase |
10870 |
0.18 |
| chr15_55557497_55557715 | 0.72 |
Mtbp |
Mdm2, transformed 3T3 cell double minute p53 binding protein |
11 |
0.65 |
| chr5_118632445_118632596 | 0.72 |
Gm43274 |
predicted gene 43274 |
11853 |
0.15 |
| chr11_95307668_95308006 | 0.71 |
Kat7 |
K(lysine) acetyltransferase 7 |
1676 |
0.25 |
| chr3_148685155_148685306 | 0.71 |
Gm43575 |
predicted gene 43575 |
32170 |
0.22 |
| chr5_17828791_17828960 | 0.70 |
Cd36 |
CD36 molecule |
6821 |
0.31 |
| chr3_121903373_121903524 | 0.70 |
Gm42593 |
predicted gene 42593 |
40606 |
0.12 |
| chr17_33523727_33524084 | 0.69 |
Adamts10 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 10 |
299 |
0.88 |
| chr4_108071243_108071567 | 0.68 |
Scp2 |
sterol carrier protein 2, liver |
14 |
0.97 |
| chr11_53519330_53520879 | 0.68 |
Septin8 |
septin 8 |
177 |
0.9 |
| chr19_10059900_10060106 | 0.67 |
Fads3 |
fatty acid desaturase 3 |
3751 |
0.16 |
| chrX_109013740_109014058 | 0.64 |
Hmgn5 |
high-mobility group nucleosome binding domain 5 |
495 |
0.74 |
| chr11_61957517_61957822 | 0.62 |
Specc1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
854 |
0.5 |
| chr10_70109912_70110290 | 0.62 |
Ccdc6 |
coiled-coil domain containing 6 |
12980 |
0.25 |
| chr6_15720466_15721441 | 0.62 |
Mdfic |
MyoD family inhibitor domain containing |
134 |
0.98 |
| chr16_17124641_17125577 | 0.62 |
2610318N02Rik |
RIKEN cDNA 2610318N02 gene |
3 |
0.87 |
| chr3_90695741_90695892 | 0.61 |
S100a9 |
S100 calcium binding protein A9 (calgranulin B) |
95 |
0.94 |
| chr7_28917363_28917885 | 0.61 |
Actn4 |
actinin alpha 4 |
1597 |
0.22 |
| chrX_48169964_48170296 | 0.59 |
Sash3 |
SAM and SH3 domain containing 3 |
23694 |
0.15 |
| chr5_30915567_30916466 | 0.59 |
Emilin1 |
elastin microfibril interfacer 1 |
2277 |
0.13 |
| chr19_40536565_40536862 | 0.59 |
Aldh18a1 |
aldehyde dehydrogenase 18 family, member A1 |
18028 |
0.13 |
| chr2_109935905_109936056 | 0.58 |
Lgr4 |
leucine-rich repeat-containing G protein-coupled receptor 4 |
18333 |
0.17 |
| chr17_46782076_46782523 | 0.57 |
Rpl7l1 |
ribosomal protein L7-like 1 |
338 |
0.8 |
| chr1_163358255_163358406 | 0.57 |
Gm24940 |
predicted gene, 24940 |
43953 |
0.11 |
| chr5_115155290_115155651 | 0.55 |
Mlec |
malectin |
2709 |
0.14 |
| chr5_34659775_34659970 | 0.52 |
Nop14 |
NOP14 nucleolar protein |
276 |
0.66 |
| chr4_108064233_108064422 | 0.52 |
Scp2 |
sterol carrier protein 2, liver |
7036 |
0.14 |
| chr16_85130982_85131137 | 0.52 |
Gm49226 |
predicted gene, 49226 |
17770 |
0.17 |
| chr10_128536302_128536612 | 0.52 |
Esyt1 |
extended synaptotagmin-like protein 1 |
10586 |
0.06 |
| chr15_57912344_57913100 | 0.50 |
Tbc1d31 |
TBC1 domain family, member 31 |
283 |
0.91 |
| chr4_135873755_135874279 | 0.50 |
Pnrc2 |
proline-rich nuclear receptor coactivator 2 |
167 |
0.91 |
| chr15_64303447_64303610 | 0.50 |
Asap1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
8161 |
0.19 |
| chr6_6578303_6578493 | 0.50 |
Sem1 |
SEM1, 26S proteasome complex subunit |
260 |
0.94 |
| chrX_12005409_12006002 | 0.49 |
Gm14512 |
predicted gene 14512 |
21237 |
0.22 |
| chr12_110870262_110870418 | 0.48 |
Cinp |
cyclin-dependent kinase 2 interacting protein |
7004 |
0.09 |
| chr12_110219876_110220027 | 0.48 |
Gm40576 |
predicted gene, 40576 |
16411 |
0.11 |
| chrX_96095960_96096527 | 0.48 |
Msn |
moesin |
146 |
0.97 |
| chr17_45733218_45734434 | 0.48 |
1600014C23Rik |
RIKEN cDNA 1600014C23 gene |
18 |
0.93 |
| chr7_19149283_19149909 | 0.48 |
Snrpd2 |
small nuclear ribonucleoprotein D2 |
126 |
0.69 |
| chr18_35751613_35752417 | 0.47 |
Sting1 |
stimulator of interferon response cGAMP interactor 1 |
11461 |
0.09 |
| chr8_113667153_113667345 | 0.47 |
Gm30052 |
predicted gene, 30052 |
9209 |
0.17 |
| chr2_170662503_170662712 | 0.46 |
Gm14264 |
predicted gene 14264 |
142 |
0.97 |
| chr19_46575201_46575567 | 0.46 |
Sfxn2 |
sideroflexin 2 |
2017 |
0.22 |
| chr6_116264345_116264698 | 0.46 |
Zfand4 |
zinc finger, AN1-type domain 4 |
256 |
0.64 |
| chr6_126939455_126939771 | 0.46 |
Rad51ap1 |
RAD51 associated protein 1 |
26 |
0.83 |
| chr13_23554912_23556285 | 0.46 |
H1f3 |
H1.3 linker histone, cluster member |
2336 |
0.07 |
| chr3_98013071_98014300 | 0.45 |
Notch2 |
notch 2 |
147 |
0.96 |
| chr17_56039992_56040714 | 0.45 |
Chaf1a |
chromatin assembly factor 1, subunit A (p150) |
86 |
0.93 |
| chrX_70476781_70477629 | 0.45 |
Tmem185a |
transmembrane protein 185A |
25 |
0.95 |
| chr14_55824498_55825973 | 0.45 |
Nfatc4 |
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
198 |
0.87 |
| chr3_129878743_129879190 | 0.45 |
Pla2g12a |
phospholipase A2, group XIIA |
152 |
0.94 |
| chr17_34982022_34982459 | 0.44 |
Lsm2 |
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
31 |
0.82 |
| chr12_103860813_103860964 | 0.44 |
Serpina1a |
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
2663 |
0.15 |
| chr7_79391983_79392204 | 0.44 |
Fanci |
Fanconi anemia, complementation group I |
164 |
0.93 |
| chr7_26055182_26055442 | 0.44 |
Cyp2b13 |
cytochrome P450, family 2, subfamily b, polypeptide 13 |
6185 |
0.14 |
| chr10_21160919_21161123 | 0.44 |
Myb |
myeloblastosis oncogene |
37 |
0.97 |
| chr7_27337516_27337823 | 0.44 |
Ltbp4 |
latent transforming growth factor beta binding protein 4 |
23 |
0.96 |
| chr9_62341481_62341954 | 0.43 |
Anp32a |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
185 |
0.95 |
| chr5_33652237_33652883 | 0.43 |
Slbp |
stem-loop binding protein |
14 |
0.87 |
| chr4_86611505_86611988 | 0.43 |
Haus6 |
HAUS augmin-like complex, subunit 6 |
273 |
0.88 |
| chr1_71102176_71102584 | 0.43 |
Bard1 |
BRCA1 associated RING domain 1 |
766 |
0.73 |
| chr8_111300149_111300519 | 0.42 |
Rfwd3 |
ring finger and WD repeat domain 3 |
112 |
0.95 |
| chr10_12964189_12964763 | 0.42 |
Stx11 |
syntaxin 11 |
178 |
0.95 |
| chr15_68362505_68362972 | 0.42 |
Gm20732 |
predicted gene 20732 |
438 |
0.76 |
| chr7_114275561_114276736 | 0.42 |
Psma1 |
proteasome subunit alpha 1 |
30 |
0.98 |
| chr3_116507746_116508585 | 0.41 |
Rtca |
RNA 3'-terminal phosphate cyclase |
32 |
0.95 |
| chr16_17619247_17619669 | 0.40 |
Smpd4 |
sphingomyelin phosphodiesterase 4 |
59 |
0.95 |
| chrX_7840897_7841392 | 0.40 |
Otud5 |
OTU domain containing 5 |
220 |
0.85 |
| chr2_34914173_34914413 | 0.40 |
Phf19 |
PHD finger protein 19 |
267 |
0.8 |
| chr10_128635554_128635990 | 0.39 |
Ikzf4 |
IKAROS family zinc finger 4 |
8876 |
0.07 |
| chr7_92743280_92743472 | 0.38 |
Rab30 |
RAB30, member RAS oncogene family |
1709 |
0.23 |
| chr9_72491893_72492093 | 0.38 |
Tex9 |
testis expressed gene 9 |
32 |
0.95 |
| chr2_93460841_93461064 | 0.38 |
Cd82 |
CD82 antigen |
960 |
0.54 |
| chrX_159255140_159255811 | 0.37 |
Rps6ka3 |
ribosomal protein S6 kinase polypeptide 3 |
307 |
0.93 |
| chr15_83122033_83122905 | 0.37 |
Rrp7a |
ribosomal RNA processing 7 homolog A (S. cerevisiae) |
282 |
0.84 |
| chr9_70275267_70275431 | 0.37 |
Myo1e |
myosin IE |
67981 |
0.1 |
| chr11_26089100_26089281 | 0.36 |
5730522E02Rik |
RIKEN cDNA 5730522E02 gene |
7590 |
0.31 |
| chr5_135107788_135108074 | 0.36 |
Mlxipl |
MLX interacting protein-like |
1013 |
0.35 |
| chr4_84523831_84524181 | 0.35 |
Bnc2 |
basonuclin 2 |
313 |
0.94 |
| chr6_136906718_136906869 | 0.35 |
Erp27 |
endoplasmic reticulum protein 27 |
15347 |
0.11 |
| chr18_35740072_35740223 | 0.35 |
Sting1 |
stimulator of interferon response cGAMP interactor 1 |
58 |
0.95 |
| chr11_98149808_98150661 | 0.34 |
Fbxl20 |
F-box and leucine-rich repeat protein 20 |
169 |
0.92 |
| chr2_26504034_26504478 | 0.33 |
Notch1 |
notch 1 |
432 |
0.73 |
| chr11_114724912_114725111 | 0.33 |
Dnaic2 |
dynein, axonemal, intermediate chain 2 |
2397 |
0.24 |
| chr17_25792314_25793132 | 0.33 |
Fam173a |
family with sequence similarity 173, member A |
75 |
0.88 |
| chr14_66977102_66977253 | 0.33 |
Gm24258 |
predicted gene, 24258 |
6725 |
0.14 |
| chr2_132252313_132253101 | 0.33 |
Pcna |
proliferating cell nuclear antigen |
105 |
0.89 |
| chr10_127677215_127677476 | 0.33 |
Nemp1 |
nuclear envelope integral membrane protein 1 |
279 |
0.67 |
| chr10_128015066_128015273 | 0.33 |
Prim1 |
DNA primase, p49 subunit |
1 |
0.94 |
| chr6_113717739_113717890 | 0.33 |
Ghrl |
ghrelin |
122 |
0.85 |
| chr5_146963438_146963654 | 0.33 |
Mtif3 |
mitochondrial translational initiation factor 3 |
197 |
0.94 |
| chr5_122820802_122821141 | 0.32 |
Anapc5 |
anaphase-promoting complex subunit 5 |
47 |
0.97 |
| chr1_136174103_136174692 | 0.32 |
Kif21b |
kinesin family member 21B |
5083 |
0.13 |
| chr2_112262662_112262916 | 0.32 |
Nop10 |
NOP10 ribonucleoprotein |
863 |
0.42 |
| chr3_36612816_36613750 | 0.32 |
Bbs7 |
Bardet-Biedl syndrome 7 (human) |
2 |
0.97 |
| chr17_35236140_35236368 | 0.31 |
Atp6v1g2 |
ATPase, H+ transporting, lysosomal V1 subunit G2 |
308 |
0.49 |
| chr7_102065670_102065844 | 0.31 |
Xndc1 |
Xrcc1 N-terminal domain containing 1 |
1 |
0.45 |
| chr14_99100247_99100434 | 0.30 |
Pibf1 |
progesterone immunomodulatory binding factor 1 |
382 |
0.6 |
| chr10_43310852_43311026 | 0.30 |
Gm16546 |
predicted gene 16546 |
8421 |
0.19 |
| chr6_38551798_38552474 | 0.30 |
Luc7l2 |
LUC7-like 2 (S. cerevisiae) |
293 |
0.87 |
| chr4_43000389_43000942 | 0.29 |
Vcp |
valosin containing protein |
158 |
0.91 |
| chr12_99561310_99561588 | 0.29 |
Foxn3 |
forkhead box N3 |
2059 |
0.21 |
| chr19_46502635_46503186 | 0.29 |
Trim8 |
tripartite motif-containing 8 |
1208 |
0.41 |
| chr14_70470911_70471105 | 0.29 |
Phyhip |
phytanoyl-CoA hydroxylase interacting protein |
12485 |
0.09 |
| chr4_3342703_3342923 | 0.28 |
Gm11786 |
predicted gene 11786 |
15638 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.8 | 2.4 | GO:0044337 | canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.8 | 3.9 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.6 | 4.5 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.5 | 2.6 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.5 | 3.7 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.4 | 1.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.3 | 1.0 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.3 | 1.7 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.3 | 1.4 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 0.7 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.2 | 4.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 | 2.6 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.2 | 0.9 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.2 | 0.6 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.2 | 0.6 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.2 | 1.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.0 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.2 | 0.7 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.2 | 0.9 | GO:0072711 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.2 | 0.5 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.2 | 0.7 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.2 | 0.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 1.1 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.2 | 0.5 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.2 | 0.5 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.2 | 0.3 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.2 | 0.5 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 0.8 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 | 0.7 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.1 | 0.4 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.1 | 0.3 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.1 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.4 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) |
| 0.1 | 1.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.3 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 2.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.3 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 2.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 0.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.3 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.1 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.6 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.3 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.1 | GO:0009757 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.1 | 0.2 | GO:0042505 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.1 | 1.5 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.1 | 0.3 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.1 | 0.4 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.1 | 0.6 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.6 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.8 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.4 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 1.6 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 1.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 1.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.0 | 0.6 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.9 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.1 | GO:0019042 | viral latency(GO:0019042) |
| 0.0 | 0.3 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.0 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.0 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.4 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.0 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 5.6 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.2 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.0 | GO:0090169 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.0 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.0 | 1.4 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.3 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.0 | GO:0060374 | mast cell differentiation(GO:0060374) |
| 0.0 | 0.1 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.0 | 0.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.0 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.0 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.0 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 1.8 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.1 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.0 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.2 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.0 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 2.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.0 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.0 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.7 | 5.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.4 | 2.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.4 | 1.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.3 | 0.8 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.2 | 2.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.2 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 2.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 5.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.2 | 1.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.7 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.7 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 1.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.4 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 2.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 4.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.6 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.5 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.2 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.3 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 7.7 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.9 | 2.7 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.7 | 3.6 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.7 | 2.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.6 | 5.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 1.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.3 | 0.9 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.3 | 2.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 0.6 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 0.9 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 1.0 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 0.5 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 2.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.8 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 3.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.3 | GO:0043338 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.1 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 1.0 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.4 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 1.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 8.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0034560 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) bidentate ribonuclease III activity(GO:0016443) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 1.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.0 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:1990190 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 2.3 | GO:0044389 | ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 10.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 1.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 6.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 2.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 2.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |