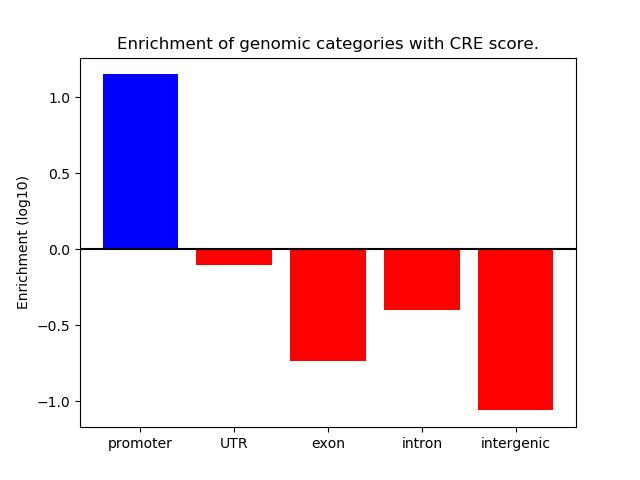
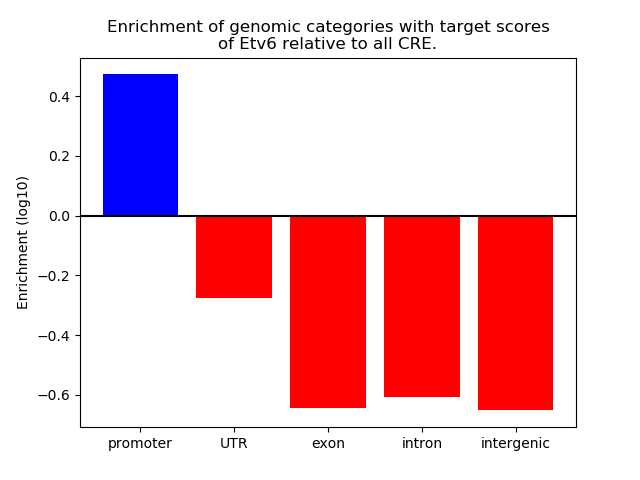
Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
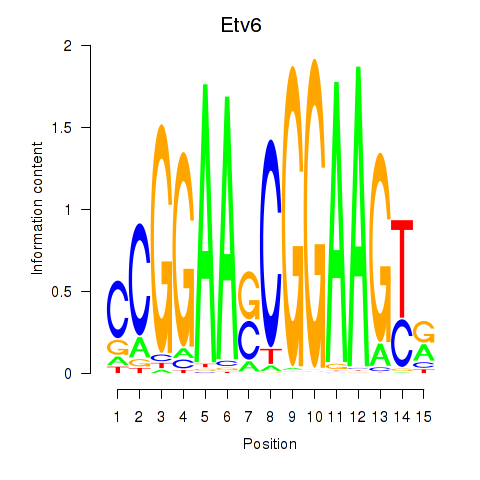
Results for Etv6
Z-value: 2.41
Transcription factors associated with Etv6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Etv6
|
ENSMUSG00000030199.10 | ets variant 6 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_134222360_134222511 | Etv6 | 11165 | 0.166117 | -0.48 | 2.0e-04 | Click! |
| chr6_134222590_134222917 | Etv6 | 10847 | 0.166630 | -0.40 | 2.2e-03 | Click! |
| chr6_134262051_134262202 | Etv6 | 14561 | 0.161313 | -0.40 | 2.2e-03 | Click! |
| chr6_134033112_134033263 | Etv6 | 2513 | 0.295838 | -0.33 | 1.3e-02 | Click! |
| chr6_134256500_134256651 | Etv6 | 9010 | 0.168388 | -0.33 | 1.5e-02 | Click! |
Activity of the Etv6 motif across conditions
Conditions sorted by the z-value of the Etv6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
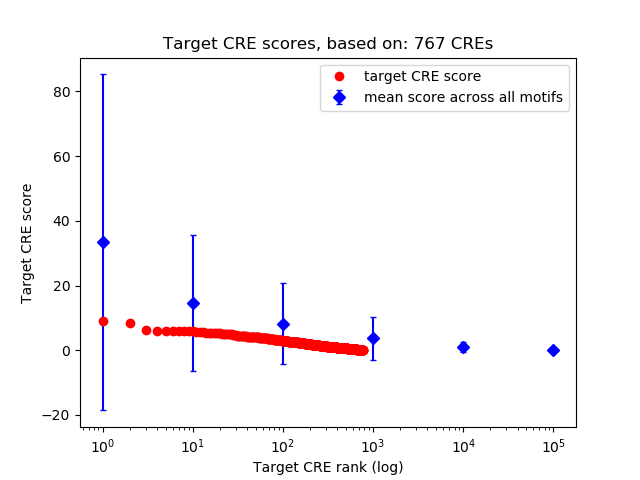
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_78861495_78861819 | 8.98 |
Atp6v1d |
ATPase, H+ transporting, lysosomal V1 subunit D |
19 |
0.67 |
| chr18_62548232_62549066 | 8.42 |
Fbxo38 |
F-box protein 38 |
35 |
0.78 |
| chr17_56405416_56405574 | 6.39 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
8935 |
0.13 |
| chr14_55671842_55672007 | 6.07 |
Gmpr2 |
guanosine monophosphate reductase 2 |
17 |
0.5 |
| chr7_105744333_105744535 | 6.06 |
Taf10 |
TATA-box binding protein associated factor 10 |
73 |
0.93 |
| chr10_13008292_13008475 | 5.97 |
Sf3b5 |
splicing factor 3b, subunit 5 |
3040 |
0.25 |
| chr8_107056570_107056952 | 5.97 |
Cog8 |
component of oligomeric golgi complex 8 |
72 |
0.55 |
| chr12_37241745_37241944 | 5.96 |
Agmo |
alkylglycerol monooxygenase |
16 |
0.99 |
| chr15_5185671_5186006 | 5.85 |
Ttc33 |
tetratricopeptide repeat domain 33 |
111 |
0.49 |
| chr6_7690583_7690734 | 5.80 |
Asns |
asparagine synthetase |
1564 |
0.43 |
| chr11_117986459_117986961 | 5.61 |
Pgs1 |
phosphatidylglycerophosphate synthase 1 |
175 |
0.93 |
| chr7_80947183_80947638 | 5.60 |
Sec11a |
SEC11 homolog A, signal peptidase complex subunit |
101 |
0.94 |
| chr2_11778402_11778601 | 5.56 |
Ankrd16 |
ankyrin repeat domain 16 |
10 |
0.95 |
| chr6_120363997_120364179 | 5.45 |
Kdm5a |
lysine (K)-specific demethylase 5A |
36 |
0.78 |
| chr12_55155228_55155411 | 5.39 |
Srp54b |
signal recognition particle 54B |
51 |
0.85 |
| chr11_3451901_3452515 | 5.36 |
Rnf185 |
ring finger protein 185 |
126 |
0.53 |
| chr1_58504889_58505222 | 5.28 |
Orc2 |
origin recognition complex, subunit 2 |
54 |
0.54 |
| chr4_124802399_124802592 | 5.26 |
Mtf1 |
metal response element binding transcription factor 1 |
54 |
0.95 |
| chr2_94010712_94010907 | 5.26 |
Alkbh3 |
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
2 |
0.97 |
| chr12_55230291_55230494 | 5.20 |
Srp54c |
signal recognition particle 54C |
224 |
0.67 |
| chr2_127208202_127208406 | 5.16 |
1810024B03Rik |
RIKEN cDNA 1810024B03 gene |
24 |
0.55 |
| chr5_31202209_31202548 | 5.16 |
Zfp513 |
zinc finger protein 513 |
75 |
0.92 |
| chr12_55080390_55080577 | 5.12 |
Srp54a |
signal recognition particle 54A |
43 |
0.84 |
| chr9_19622136_19622591 | 4.96 |
Zfp317 |
zinc finger protein 317 |
59 |
0.96 |
| chr5_116024394_116024784 | 4.95 |
Prkab1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
81 |
0.95 |
| chr13_41000869_41001065 | 4.90 |
Pak1ip1 |
PAK1 interacting protein 1 |
56 |
0.67 |
| chr2_132253158_132253669 | 4.86 |
AV099323 |
expressed sequence AV099323 |
56 |
0.55 |
| chr14_26669845_26670062 | 4.67 |
9930004E17Rik |
RIKEN cDNA 9930004E17 gene |
57 |
0.46 |
| chr3_146521363_146521539 | 4.63 |
Rpf1 |
ribosome production factor 1 homolog |
22 |
0.97 |
| chr17_24617516_24617667 | 4.63 |
Tsc2 |
TSC complex subunit 2 |
3010 |
0.11 |
| chr17_36271341_36271519 | 4.51 |
Trim39 |
tripartite motif-containing 39 |
47 |
0.92 |
| chr2_152226771_152226948 | 4.48 |
Csnk2a1 |
casein kinase 2, alpha 1 polypeptide |
17 |
0.97 |
| chr14_52278918_52280079 | 4.45 |
Rab2b |
RAB2B, member RAS oncogene family |
47 |
0.51 |
| chr17_21733492_21733782 | 4.44 |
Zfp229 |
zinc finger protein 229 |
43 |
0.96 |
| chrX_169320012_169320582 | 4.42 |
Hccs |
holocytochrome c synthetase |
3 |
0.84 |
| chr18_46741720_46741894 | 4.32 |
Ap3s1 |
adaptor-related protein complex 3, sigma 1 subunit |
69 |
0.71 |
| chr17_20944797_20945472 | 4.29 |
Gm49761 |
predicted gene, 49761 |
15 |
0.68 |
| chr2_145934675_145934882 | 4.26 |
Cfap61 |
cilia and flagella associated protein 61 |
6 |
0.53 |
| chr7_109082988_109083506 | 4.24 |
Ric3 |
RIC3 acetylcholine receptor chaperone |
58 |
0.97 |
| chr4_125066497_125066766 | 4.19 |
Snip1 |
Smad nuclear interacting protein 1 |
41 |
0.93 |
| chr19_5366571_5366780 | 4.18 |
Eif1ad |
eukaryotic translation initiation factor 1A domain containing |
66 |
0.48 |
| chr11_106255719_106255906 | 4.16 |
Ftsj3 |
FtsJ RNA methyltransferase homolog 3 (E. coli) |
8 |
0.82 |
| chr14_101640358_101640529 | 4.14 |
Commd6 |
COMM domain containing 6 |
5 |
0.97 |
| chr19_8871301_8872099 | 4.14 |
Ubxn1 |
UBX domain protein 1 |
40 |
0.9 |
| chr2_130667648_130667858 | 4.12 |
Itpa |
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
52 |
0.95 |
| chr11_94653662_94653850 | 4.12 |
Mrpl27 |
mitochondrial ribosomal protein L27 |
11 |
0.55 |
| chr2_132686675_132687262 | 4.09 |
Shld1 |
shieldin complex subunit 1 |
37 |
0.96 |
| chr2_37443118_37443593 | 4.08 |
Rabgap1 |
RAB GTPase activating protein 1 |
59 |
0.54 |
| chr14_52197103_52197330 | 4.07 |
Supt16 |
SPT16, facilitates chromatin remodeling subunit |
200 |
0.58 |
| chr5_36830527_36830904 | 4.04 |
Man2b2 |
mannosidase 2, alpha B2 |
62 |
0.97 |
| chr11_44529187_44529856 | 4.02 |
Rnf145 |
ring finger protein 145 |
9216 |
0.18 |
| chr17_35969652_35970052 | 3.98 |
Abcf1 |
ATP-binding cassette, sub-family F (GCN20), member 1 |
91 |
0.89 |
| chr1_179546195_179546570 | 3.94 |
Cnst |
consortin, connexin sorting protein |
12 |
0.6 |
| chr11_60830808_60831009 | 3.93 |
Dhrs7b |
dehydrogenase/reductase (SDR family) member 7B |
229 |
0.86 |
| chr17_5286471_5286905 | 3.92 |
Gm29050 |
predicted gene 29050 |
102075 |
0.07 |
| chr9_70421744_70422098 | 3.90 |
Ccnb2 |
cyclin B2 |
374 |
0.84 |
| chr10_128805324_128805862 | 3.90 |
Dnajc14 |
DnaJ heat shock protein family (Hsp40) member C14 |
10 |
0.93 |
| chr1_193173359_193174042 | 3.84 |
A130010J15Rik |
RIKEN cDNA A130010J15 gene |
44 |
0.95 |
| chr1_167308575_167308778 | 3.84 |
Tmco1 |
transmembrane and coiled-coil domains 1 |
6 |
0.96 |
| chr16_56037403_56037620 | 3.80 |
Trmt10c |
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
308 |
0.79 |
| chr4_137388371_137388553 | 3.77 |
2810405F17Rik |
RIKEN cDNA 2810405F17 gene |
80 |
0.95 |
| chr3_88424832_88425399 | 3.71 |
Slc25a44 |
solute carrier family 25, member 44 |
3 |
0.93 |
| chr2_30967430_30968192 | 3.68 |
Tor1a |
torsin family 1, member A (torsin A) |
61 |
0.96 |
| chr6_113046036_113046851 | 3.67 |
Thumpd3 |
THUMP domain containing 3 |
15 |
0.8 |
| chr15_80097023_80097326 | 3.65 |
Syngr1 |
synaptogyrin 1 |
696 |
0.49 |
| chr8_105297551_105298218 | 3.65 |
E2f4 |
E2F transcription factor 4 |
167 |
0.83 |
| chr7_127471210_127471989 | 3.63 |
Prr14 |
proline rich 14 |
109 |
0.89 |
| chr15_10485915_10486089 | 3.60 |
Rad1 |
RAD1 checkpoint DNA exonuclease |
16 |
0.52 |
| chr15_12824603_12824975 | 3.60 |
Drosha |
drosha, ribonuclease type III |
26 |
0.63 |
| chr4_115850014_115850176 | 3.57 |
Mknk1 |
MAP kinase-interacting serine/threonine kinase 1 |
6937 |
0.13 |
| chr6_125009531_125009725 | 3.57 |
Zfp384 |
zinc finger protein 384 |
41 |
0.94 |
| chr6_136828152_136828539 | 3.55 |
Wbp11 |
WW domain binding protein 11 |
112 |
0.76 |
| chr2_39007642_39008238 | 3.52 |
Arpc5l |
actin related protein 2/3 complex, subunit 5-like |
136 |
0.91 |
| chr1_36243812_36244462 | 3.51 |
Uggt1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
24 |
0.97 |
| chr6_17797574_17797931 | 3.49 |
Gm26738 |
predicted gene, 26738 |
37433 |
0.13 |
| chr15_58933660_58933845 | 3.47 |
Ndufb9 |
NADH:ubiquinone oxidoreductase subunit B9 |
4 |
0.49 |
| chr3_157534009_157535172 | 3.47 |
Zranb2 |
zinc finger, RAN-binding domain containing 2 |
165 |
0.81 |
| chr2_121473954_121474145 | 3.39 |
Mfap1b |
microfibrillar-associated protein 1B |
18 |
0.94 |
| chr9_65032072_65032479 | 3.32 |
Dpp8 |
dipeptidylpeptidase 8 |
139 |
0.95 |
| chr1_184441391_184441542 | 3.31 |
2900092O11Rik |
RIKEN cDNA 2900092O11 gene |
1514 |
0.45 |
| chr4_53826161_53826553 | 3.28 |
Tmem38b |
transmembrane protein 38B |
300 |
0.89 |
| chr15_79670716_79670928 | 3.28 |
Tomm22 |
translocase of outer mitochondrial membrane 22 |
39 |
0.94 |
| chr12_100159598_100160070 | 3.26 |
Nrde2 |
nrde-2 necessary for RNA interference, domain containing |
181 |
0.93 |
| chr19_45006351_45006736 | 3.26 |
Twnk |
twinkle mtDNA helicase |
15 |
0.57 |
| chr2_130424198_130424383 | 3.25 |
Pced1a |
PC-esterase domain containing 1A |
9 |
0.51 |
| chr15_30610696_30610847 | 3.20 |
Gm24487 |
predicted gene, 24487 |
9257 |
0.18 |
| chr2_181319482_181319804 | 3.19 |
Rtel1 |
regulator of telomere elongation helicase 1 |
96 |
0.93 |
| chrX_100625656_100626100 | 3.19 |
Pdzd11 |
PDZ domain containing 11 |
57 |
0.61 |
| chr8_72570981_72571279 | 3.17 |
Smim7 |
small integral membrane protein 7 |
42 |
0.93 |
| chr7_127421975_127422168 | 3.15 |
Zfp688 |
zinc finger protein 688 |
3 |
0.93 |
| chr2_26389102_26389724 | 3.10 |
Pmpca |
peptidase (mitochondrial processing) alpha |
57 |
0.53 |
| chr13_21362676_21363275 | 3.04 |
Zscan12 |
zinc finger and SCAN domain containing 12 |
143 |
0.9 |
| chr1_45794842_45795231 | 3.02 |
Wdr75 |
WD repeat domain 75 |
130 |
0.95 |
| chr9_20385074_20385289 | 3.01 |
Zfp560 |
zinc finger protein 560 |
4 |
0.52 |
| chr15_8109173_8109361 | 3.01 |
Nup155 |
nucleoporin 155 |
6 |
0.98 |
| chr17_87025276_87025630 | 3.01 |
Pigf |
phosphatidylinositol glycan anchor biosynthesis, class F |
47 |
0.55 |
| chr13_36117784_36118119 | 3.00 |
Fars2 |
phenylalanine-tRNA synthetase 2 (mitochondrial) |
71 |
0.86 |
| chr10_83359933_83360353 | 2.99 |
D10Wsu102e |
DNA segment, Chr 10, Wayne State University 102, expressed |
78 |
0.97 |
| chr12_85110707_85110908 | 2.98 |
Dlst |
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
26 |
0.51 |
| chr12_85374188_85374785 | 2.97 |
Tmed10 |
transmembrane p24 trafficking protein 10 |
204 |
0.66 |
| chr17_40934490_40934852 | 2.96 |
Mmut |
methylmalonyl-Coenzyme A mutase |
14 |
0.61 |
| chr11_95011751_95011961 | 2.94 |
Samd14 |
sterile alpha motif domain containing 14 |
1575 |
0.24 |
| chr7_126474470_126475241 | 2.94 |
Sh2b1 |
SH2B adaptor protein 1 |
31 |
0.95 |
| chr6_35539763_35540120 | 2.93 |
Mtpn |
myotrophin |
53 |
0.98 |
| chr1_167392688_167392913 | 2.86 |
Mgst3 |
microsomal glutathione S-transferase 3 |
1041 |
0.48 |
| chr2_29935294_29935511 | 2.83 |
Gle1 |
GLE1 RNA export mediator (yeast) |
24 |
0.96 |
| chr11_67588922_67589098 | 2.80 |
Gas7 |
growth arrest specific 7 |
2309 |
0.33 |
| chr9_37348283_37348681 | 2.79 |
Ccdc15 |
coiled-coil domain containing 15 |
50 |
0.95 |
| chr4_31963680_31964247 | 2.76 |
Map3k7 |
mitogen-activated protein kinase kinase kinase 7 |
134 |
0.97 |
| chr1_33669479_33669900 | 2.75 |
Prim2 |
DNA primase, p58 subunit |
106 |
0.53 |
| chr2_140066689_140066983 | 2.75 |
Tasp1 |
taspase, threonine aspartase 1 |
31 |
0.98 |
| chr6_120822279_120822990 | 2.75 |
Atp6v1e1 |
ATPase, H+ transporting, lysosomal V1 subunit E1 |
51 |
0.97 |
| chr10_127290569_127290765 | 2.72 |
Gm4189 |
predicted gene 4189 |
57 |
0.54 |
| chr2_11332597_11332748 | 2.72 |
Gm37851 |
predicted gene, 37851 |
153 |
0.92 |
| chr11_29526267_29526597 | 2.70 |
Mtif2 |
mitochondrial translational initiation factor 2 |
4 |
0.96 |
| chr9_20952766_20953077 | 2.69 |
Dnmt1 |
DNA methyltransferase (cytosine-5) 1 |
58 |
0.94 |
| chr4_40948174_40948980 | 2.69 |
Chmp5 |
charged multivesicular body protein 5 |
170 |
0.63 |
| chr12_84970815_84970994 | 2.68 |
Arel1 |
apoptosis resistant E3 ubiquitin protein ligase 1 |
4 |
0.49 |
| chr11_87737188_87737626 | 2.68 |
Supt4a |
SPT4A, DSIF elongation factor subunit |
145 |
0.91 |
| chr9_79759713_79759940 | 2.66 |
Cox7a2 |
cytochrome c oxidase subunit 7A2 |
52 |
0.97 |
| chr8_124520582_124520885 | 2.66 |
Cog2 |
component of oligomeric golgi complex 2 |
34 |
0.98 |
| chr11_5485632_5485992 | 2.65 |
Gm11962 |
predicted gene 11962 |
9634 |
0.14 |
| chr11_83298909_83299212 | 2.65 |
Ap2b1 |
adaptor-related protein complex 2, beta 1 subunit |
36 |
0.54 |
| chr7_56049865_56050780 | 2.64 |
Herc2 |
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
67 |
0.81 |
| chr4_150568722_150569341 | 2.62 |
Gm13091 |
predicted gene 13091 |
487 |
0.51 |
| chr1_161968669_161969306 | 2.61 |
4930558K02Rik |
RIKEN cDNA 4930558K02 gene |
161 |
0.39 |
| chr13_17993258_17993531 | 2.60 |
Yae1d1 |
Yae1 domain containing 1 |
45 |
0.92 |
| chr15_34443090_34443386 | 2.59 |
Rpl30 |
ribosomal protein L30 |
16 |
0.77 |
| chr3_95871394_95871675 | 2.59 |
C920021L13Rik |
RIKEN cDNA C920021L13 gene |
3 |
0.49 |
| chr11_86757222_86757692 | 2.54 |
Cltc |
clathrin, heavy polypeptide (Hc) |
44 |
0.98 |
| chr15_51865432_51865773 | 2.54 |
Eif3h |
eukaryotic translation initiation factor 3, subunit H |
79 |
0.97 |
| chr6_149309318_149309909 | 2.52 |
Resf1 |
retroelement silencing factor 1 |
40 |
0.97 |
| chr11_114668310_114668676 | 2.52 |
Rpl38 |
ribosomal protein L38 |
31 |
0.97 |
| chr11_58330284_58330451 | 2.50 |
Zfp672 |
zinc finger protein 672 |
28 |
0.83 |
| chr1_57969978_57970355 | 2.50 |
Kctd18 |
potassium channel tetramerisation domain containing 18 |
38 |
0.98 |
| chr18_74267773_74268322 | 2.50 |
Mbd1 |
methyl-CpG binding domain protein 1 |
128 |
0.95 |
| chr5_145201828_145202036 | 2.49 |
Zkscan14 |
zinc finger with KRAB and SCAN domains 14 |
64 |
0.94 |
| chr16_21332982_21333472 | 2.48 |
Magef1 |
melanoma antigen family F, 1 |
98 |
0.97 |
| chr2_12419340_12419682 | 2.47 |
Mindy3 |
MINDY lysine 48 deubiquitinase 3 |
41 |
0.97 |
| chr19_32485724_32486055 | 2.44 |
Minpp1 |
multiple inositol polyphosphate histidine phosphatase 1 |
34 |
0.49 |
| chr10_14704849_14705669 | 2.44 |
Vta1 |
vesicle (multivesicular body) trafficking 1 |
200 |
0.64 |
| chrX_94212852_94213030 | 2.43 |
Eif2s3x |
eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked |
79 |
0.97 |
| chrX_107816252_107816458 | 2.42 |
2610002M06Rik |
RIKEN cDNA 2610002M06 gene |
21 |
0.61 |
| chr17_56764467_56764869 | 2.41 |
Dus3l |
dihydrouridine synthase 3-like (S. cerevisiae) |
89 |
0.94 |
| chr7_66079145_66079708 | 2.40 |
Gm45081 |
predicted gene 45081 |
47 |
0.72 |
| chr17_26662965_26663282 | 2.39 |
Atp6v0e |
ATPase, H+ transporting, lysosomal V0 subunit E |
108 |
0.95 |
| chr11_20914053_20914257 | 2.39 |
Gm23681 |
predicted gene, 23681 |
23546 |
0.17 |
| chr19_8770894_8771102 | 2.38 |
Tmem223 |
transmembrane protein 223 |
2 |
0.87 |
| chr8_10854830_10855021 | 2.35 |
Gm32540 |
predicted gene, 32540 |
11261 |
0.12 |
| chr2_153425549_153426538 | 2.34 |
Gm14472 |
predicted gene 14472 |
12194 |
0.16 |
| chr18_75000413_75000592 | 2.31 |
Rpl17 |
ribosomal protein L17 |
25 |
0.86 |
| chr11_120457992_120458200 | 2.30 |
Ccdc137 |
coiled-coil domain containing 137 |
19 |
0.49 |
| chrX_101287598_101287912 | 2.28 |
Med12 |
mediator complex subunit 12 |
1731 |
0.2 |
| chr6_113378048_113378624 | 2.28 |
Arpc4 |
actin related protein 2/3 complex, subunit 4 |
143 |
0.76 |
| chr5_142895139_142895334 | 2.27 |
Fbxl18 |
F-box and leucine-rich repeat protein 18 |
2 |
0.97 |
| chr10_22730737_22731547 | 2.26 |
AC166256.1 |
novel protein |
95 |
0.67 |
| chr14_51884584_51884965 | 2.26 |
Mettl17 |
methyltransferase like 17 |
68 |
0.94 |
| chr3_95855485_95855981 | 2.25 |
Prpf3 |
pre-mRNA processing factor 3 |
20 |
0.74 |
| chrX_140956364_140956986 | 2.23 |
Psmd10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
14 |
0.76 |
| chr18_30272681_30273169 | 2.22 |
Pik3c3 |
phosphatidylinositol 3-kinase catalytic subunit type 3 |
2 |
0.98 |
| chr11_120624901_120625106 | 2.20 |
Sirt7 |
sirtuin 7 |
4 |
0.6 |
| chr10_78247305_78247456 | 2.20 |
Trappc10 |
trafficking protein particle complex 10 |
2739 |
0.15 |
| chr3_94886609_94887089 | 2.17 |
Psmb4 |
proteasome (prosome, macropain) subunit, beta type 4 |
13 |
0.96 |
| chr10_58255270_58255969 | 2.16 |
Gcc2 |
GRIP and coiled-coil domain containing 2 |
80 |
0.96 |
| chr9_20520924_20521753 | 2.14 |
Zfp266 |
zinc finger protein 266 |
70 |
0.95 |
| chr18_36783049_36783284 | 2.13 |
Hars |
histidyl-tRNA synthetase |
39 |
0.51 |
| chr11_16508124_16508749 | 2.12 |
Sec61g |
SEC61, gamma subunit |
48 |
0.98 |
| chr8_26015514_26015770 | 2.12 |
Fnta |
farnesyltransferase, CAAX box, alpha |
8 |
0.96 |
| chr17_86301214_86301583 | 2.12 |
2010106C02Rik |
RIKEN cDNA 2010106C02 gene |
14220 |
0.27 |
| chr6_118419131_118419637 | 2.11 |
Bms1 |
BMS1, ribosome biogenesis factor |
20 |
0.97 |
| chr6_88518714_88519337 | 2.10 |
Gm44430 |
predicted gene, 44430 |
12 |
0.61 |
| chr11_77793211_77793566 | 2.10 |
Gm10277 |
predicted gene 10277 |
5641 |
0.16 |
| chr10_111972464_111972783 | 2.09 |
Krr1 |
KRR1, small subunit (SSU) processome component, homolog (yeast) |
41 |
0.97 |
| chr13_4190910_4191302 | 2.09 |
Akr1c13 |
aldo-keto reductase family 1, member C13 |
44 |
0.96 |
| chr6_128424839_128425024 | 2.08 |
Itfg2 |
integrin alpha FG-GAP repeat containing 2 |
0 |
0.94 |
| chr7_127232832_127233741 | 2.07 |
Zfp553 |
zinc finger protein 553 |
8 |
0.65 |
| chr1_106796168_106796838 | 2.06 |
Vps4b |
vacuolar protein sorting 4B |
74 |
0.97 |
| chr10_30600606_30601328 | 2.06 |
Trmt11 |
tRNA methyltransferase 11 |
218 |
0.91 |
| chr2_10079825_10080668 | 2.05 |
Atp5c1 |
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
70 |
0.8 |
| chr6_128754257_128754598 | 2.04 |
Gm43907 |
predicted gene, 43907 |
9783 |
0.11 |
| chr4_136886113_136886264 | 2.03 |
C1qb |
complement component 1, q subcomponent, beta polypeptide |
1 |
0.97 |
| chr1_133610228_133610578 | 2.02 |
Snrpe |
small nuclear ribonucleoprotein E |
112 |
0.95 |
| chr19_38223870_38224599 | 2.01 |
Fra10ac1 |
FRA10AC1 homolog (human) |
78 |
0.97 |
| chr7_12909991_12910417 | 2.01 |
Rps5 |
ribosomal protein S5 |
12086 |
0.07 |
| chr13_67525535_67526312 | 1.96 |
Zfp87 |
zinc finger protein 87 |
198 |
0.51 |
| chr2_152831597_152831827 | 1.95 |
Bcl2l1 |
BCL2-like 1 |
2 |
0.97 |
| chr10_62327332_62327499 | 1.95 |
Hk1 |
hexokinase 1 |
352 |
0.86 |
| chr7_67645147_67645521 | 1.94 |
Lrrc28 |
leucine rich repeat containing 28 |
66 |
0.96 |
| chr17_33929330_33929641 | 1.94 |
Rgl2 |
ral guanine nucleotide dissociation stimulator-like 2 |
58 |
0.88 |
| chr5_24586600_24586907 | 1.93 |
Chpf2 |
chondroitin polymerizing factor 2 |
3 |
0.95 |
| chr16_62786657_62787006 | 1.93 |
Nsun3 |
NOL1/NOP2/Sun domain family member 3 |
13 |
0.97 |
| chr11_77078150_77078604 | 1.91 |
Nsrp1 |
nuclear speckle regulatory protein 1 |
58 |
0.69 |
| chr10_128547748_128547947 | 1.90 |
Zc3h10 |
zinc finger CCCH type containing 10 |
73 |
0.89 |
| chr9_107562836_107563301 | 1.89 |
Tusc2 |
tumor suppressor 2, mitochondrial calcium regulator |
187 |
0.8 |
| chr17_35979236_35979423 | 1.89 |
Prr3 |
proline-rich polypeptide 3 |
30 |
0.86 |
| chr4_82859084_82859933 | 1.88 |
Zdhhc21 |
zinc finger, DHHC domain containing 21 |
46 |
0.98 |
| chr1_190724564_190725420 | 1.87 |
Rps6kc1 |
ribosomal protein S6 kinase polypeptide 1 |
85471 |
0.09 |
| chr3_108256684_108257282 | 1.87 |
1700010K24Rik |
RIKEN cDNA 1700010K24 gene |
2 |
0.52 |
| chr16_91464461_91465455 | 1.87 |
A930006K02Rik |
RIKEN cDNA A930006K02 gene |
67 |
0.46 |
| chr14_70336335_70336839 | 1.86 |
Slc39a14 |
solute carrier family 39 (zinc transporter), member 14 |
2389 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 1.6 | 4.9 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 1.2 | 3.7 | GO:2000152 | regulation of ubiquitin-specific protease activity(GO:2000152) |
| 1.1 | 5.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.1 | 1.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 1.0 | 4.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 1.0 | 2.0 | GO:0019042 | viral latency(GO:0019042) |
| 1.0 | 2.9 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.9 | 2.8 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.8 | 2.5 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.8 | 2.5 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.8 | 3.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.8 | 3.9 | GO:0009146 | purine nucleoside triphosphate catabolic process(GO:0009146) |
| 0.8 | 2.3 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.8 | 3.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.8 | 3.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.8 | 3.8 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.7 | 4.3 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.7 | 2.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.7 | 2.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.7 | 2.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.7 | 2.0 | GO:0090045 | positive regulation of deacetylase activity(GO:0090045) |
| 0.6 | 1.9 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.6 | 1.9 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.6 | 5.6 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.6 | 1.7 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.6 | 1.7 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.6 | 1.7 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.5 | 1.6 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.5 | 1.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.5 | 3.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.5 | 2.7 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.5 | 1.6 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.5 | 2.6 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.5 | 6.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.5 | 1.5 | GO:0052200 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.5 | 7.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.5 | 3.0 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.5 | 2.5 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.5 | 1.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 1.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.5 | 4.2 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.5 | 1.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.5 | 5.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.5 | 0.9 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.4 | 3.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 0.9 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 1.3 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.4 | 2.5 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.4 | 1.3 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.4 | 1.6 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.4 | 1.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.4 | 5.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.4 | 1.4 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.4 | 1.4 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.3 | 1.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.3 | 1.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.3 | 8.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 1.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.3 | 1.0 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 2.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 1.0 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.3 | 0.9 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.3 | 1.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.3 | 1.9 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.3 | 1.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 2.2 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.3 | 1.2 | GO:0061724 | lipophagy(GO:0061724) |
| 0.3 | 4.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.3 | 3.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 1.2 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.3 | 1.2 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.3 | 0.9 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.3 | 1.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.3 | 1.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.3 | 0.8 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 0.8 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.3 | 0.8 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.3 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 1.5 | GO:0061620 | glycolytic process through glucose-6-phosphate(GO:0061620) |
| 0.3 | 0.8 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.2 | 2.4 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 | 3.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 3.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 0.7 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 0.7 | GO:0046075 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.2 | 1.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 1.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 6.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 1.2 | GO:0090188 | negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.2 | 2.8 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 0.8 | GO:0042524 | negative regulation of tyrosine phosphorylation of Stat5 protein(GO:0042524) |
| 0.2 | 0.8 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 3.1 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 0.8 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 | 1.9 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) |
| 0.2 | 0.7 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.2 | 1.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 0.5 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.2 | 0.9 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 1.4 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 | 0.4 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.2 | 0.5 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.2 | 6.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 0.7 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.2 | 0.5 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
| 0.2 | 2.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.2 | 1.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 0.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 5.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.2 | 1.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 0.8 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 4.9 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.2 | 0.6 | GO:0008228 | opsonization(GO:0008228) |
| 0.2 | 2.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 0.6 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.2 | 2.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.2 | 2.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 0.3 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.2 | 4.7 | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition(GO:0045841) mitotic spindle checkpoint(GO:0071174) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.1 | 0.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.9 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 3.5 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.1 | 0.4 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 1.7 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.5 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 1.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 0.4 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 1.3 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 1.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 1.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 2.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 5.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.5 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 1.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.3 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 4.5 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 1.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 5.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 1.5 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.1 | 0.4 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.5 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.6 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.5 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.5 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.1 | 2.0 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 1.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 0.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.3 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.1 | 2.3 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 1.1 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 0.5 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.4 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) regulation of metaphase/anaphase transition of cell cycle(GO:1902099) |
| 0.1 | 1.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.8 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 0.2 | GO:1903288 | regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 1.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.1 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.4 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 0.2 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.1 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 5.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 3.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 3.0 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 1.0 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 2.7 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.1 | 0.1 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.1 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.1 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.1 | 0.3 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.6 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.3 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.1 | 2.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.5 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.5 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 3.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.3 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.9 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.0 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 0.6 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 6.7 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.1 | 0.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 2.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 0.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.3 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.1 | 2.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 0.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.2 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.1 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.7 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.2 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.0 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.2 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 1.2 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 1.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.4 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.0 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.4 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:2000587 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 4.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.0 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.5 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 3.5 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 3.3 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.2 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 1.9 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0003340 | negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) |
| 0.0 | 6.2 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 2.8 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.0 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 2.2 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 1.2 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.1 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.0 | GO:2000520 | regulation of immunological synapse formation(GO:2000520) |
| 0.0 | 0.2 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.2 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.0 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.0 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.3 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.0 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.0 | GO:0042738 | biphenyl metabolic process(GO:0018879) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.0 | GO:0090224 | regulation of spindle organization(GO:0090224) |
| 0.0 | 0.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.3 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.0 | 0.0 | GO:2000977 | regulation of forebrain neuron differentiation(GO:2000977) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.0 | 0.0 | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process(GO:0009147) |
| 0.0 | 0.0 | GO:0061046 | regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.0 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.0 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.0 | GO:0033047 | regulation of mitotic sister chromatid segregation(GO:0033047) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:2000316 | regulation of T-helper 17 type immune response(GO:2000316) positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.5 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.0 | GO:0031049 | programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.0 | 0.2 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.0 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 1.9 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 2.1 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.1 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.3 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.0 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.0 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) |
| 0.0 | 0.0 | GO:1990036 | calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.0 | GO:1901525 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.0 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.0 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.0 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.0 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) |
| 0.0 | 0.8 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.0 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.0 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.2 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.9 | GO:0051168 | nuclear export(GO:0051168) |
| 0.0 | 0.1 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.0 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0000125 | PCAF complex(GO:0000125) |
| 1.3 | 2.7 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 1.3 | 3.9 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 1.3 | 8.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 1.0 | 2.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.9 | 2.8 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.9 | 2.7 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.8 | 2.5 | GO:0031523 | Myb complex(GO:0031523) |
| 0.8 | 5.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.7 | 3.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.7 | 4.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.7 | 2.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.5 | 1.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.5 | 3.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.5 | 5.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.5 | 5.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.5 | 1.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.5 | 1.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.5 | 0.9 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.5 | 3.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 3.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.4 | 4.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.4 | 4.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 2.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.4 | 1.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.4 | 1.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.4 | 1.9 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.4 | 3.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.4 | 1.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.4 | 2.8 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 1.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.3 | 2.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 3.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.3 | 7.3 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.3 | 3.5 | GO:0001527 | microfibril(GO:0001527) |
| 0.3 | 2.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 2.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 0.9 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.3 | 1.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 2.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 1.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.3 | 1.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 2.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.3 | 0.8 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.3 | 6.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 1.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 1.2 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 0.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.2 | 1.5 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 0.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 2.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 2.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 2.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 6.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 1.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 2.0 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 0.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 0.5 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 0.6 | GO:0035363 | histone locus body(GO:0035363) |
| 0.2 | 1.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 1.6 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.2 | 2.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 0.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.4 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.1 | 5.2 | GO:0030684 | preribosome(GO:0030684) |
| 0.1 | 5.6 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 4.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.4 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 1.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.4 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.1 | 0.7 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 1.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.1 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.7 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 3.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 2.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.3 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 1.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 4.7 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.1 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 1.3 | GO:0030894 | replisome(GO:0030894) |
| 0.1 | 0.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.4 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.1 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 3.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.9 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.1 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.7 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 4.8 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 9.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 38.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 24.7 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 2.8 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.0 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.0 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.0 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 1.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.8 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 3.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 5.1 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.0 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.9 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.0 | GO:0046930 | membrane attack complex(GO:0005579) pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 15.2 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 0.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0032590 | dendrite membrane(GO:0032590) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 1.7 | 5.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 1.1 | 4.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 1.1 | 5.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.9 | 3.7 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.8 | 5.9 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.8 | 2.4 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.8 | 3.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.7 | 2.2 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.7 | 2.0 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.6 | 3.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.6 | 2.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.5 | 1.6 | GO:0032357 | oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.5 | 2.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.5 | 2.0 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.5 | 2.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 1.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.5 | 1.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.5 | 1.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 4.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.4 | 1.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.4 | 3.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.4 | 2.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 2.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.4 | 3.5 | GO:0034522 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.4 | 3.9 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.4 | 1.5 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 1.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.4 | 1.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 3.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 2.8 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.4 | 4.6 | GO:0036222 | XTP diphosphatase activity(GO:0036222) |
| 0.3 | 2.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.3 | 4.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 5.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 3.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.3 | 0.9 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.3 | 2.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 3.0 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 1.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.3 | 3.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 5.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 3.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.3 | 12.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 1.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 0.8 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.2 | 1.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 1.5 | GO:0052630 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.2 | 1.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 3.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 2.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.2 | 0.9 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.2 | 0.7 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.2 | 3.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 2.1 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.2 | 4.0 | GO:0052769 | dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.2 | 6.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.2 | 1.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 1.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 1.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 3.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 0.6 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.2 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 0.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 0.6 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 5.2 | GO:0034792 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.2 | 0.3 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.2 | 4.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 1.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 2.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 1.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 0.6 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 4.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 1.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.2 | 1.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 0.5 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.2 | 0.9 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 1.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 3.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 11.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 2.1 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.1 | 0.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.7 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 5.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.4 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.1 | 1.7 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.5 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.1 | 3.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 3.6 | GO:0016751 | S-succinyltransferase activity(GO:0016751) |
| 0.1 | 0.8 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 5.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 1.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 7.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 8.0 | GO:0015405 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 1.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.2 | GO:0042281 | dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042281) |
| 0.1 | 0.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 6.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 2.1 | GO:0004495 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.1 | 0.2 | GO:1904680 | oligopeptide transmembrane transporter activity(GO:0035673) peptide transmembrane transporter activity(GO:1904680) |
| 0.1 | 1.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 1.7 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 1.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 4.6 | GO:0046921 | alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.2 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.1 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.6 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 4.3 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.1 | 0.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.3 | GO:0032404 | mismatch repair complex binding(GO:0032404) |
| 0.1 | 0.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 3.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 5.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.1 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 5.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.0 | 0.2 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 1.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 1.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 8.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.7 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 1.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.8 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 8.4 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 1.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 2.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 1.0 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 4.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.3 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 2.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.0 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 4.8 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 17.2 | GO:0044822 | poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.7 | GO:0052767 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.1 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 1.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.1 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 0.1 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 1.4 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.9 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.0 | 0.3 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.0 | 0.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.0 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 3.7 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.0 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 2.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.0 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0015928 | fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.0 | GO:0003916 | DNA topoisomerase activity(GO:0003916) DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.3 | 3.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 4.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 3.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 2.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 1.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 5.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 6.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 3.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 0.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.7 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 2.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.6 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.0 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.5 | 2.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 5.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.4 | 4.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.4 | 2.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.4 | 7.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 4.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 7.9 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.3 | 2.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 2.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 3.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.3 | 3.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 2.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.3 | 2.9 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 3.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 2.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 2.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 1.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 1.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 3.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 8.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.2 | 3.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 3.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 5.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 6.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.7 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 3.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 4.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 4.3 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 1.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 5.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 1.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 3.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.1 | 1.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.1 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.1 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 1.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.6 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.7 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.2 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 5.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.0 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | Genes involved in Post-translational protein modification |
| 0.0 | 0.0 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |