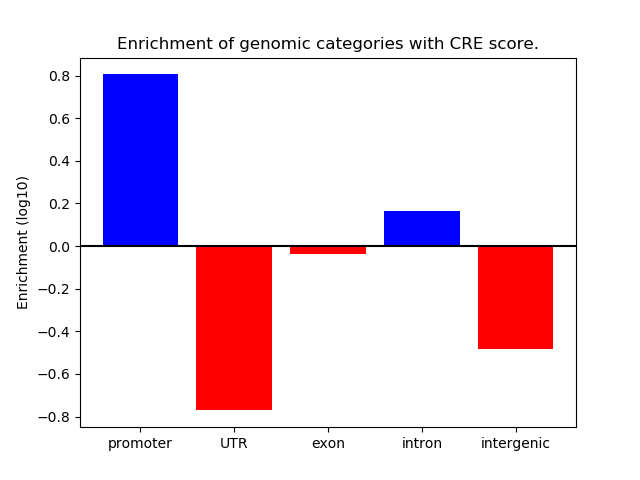
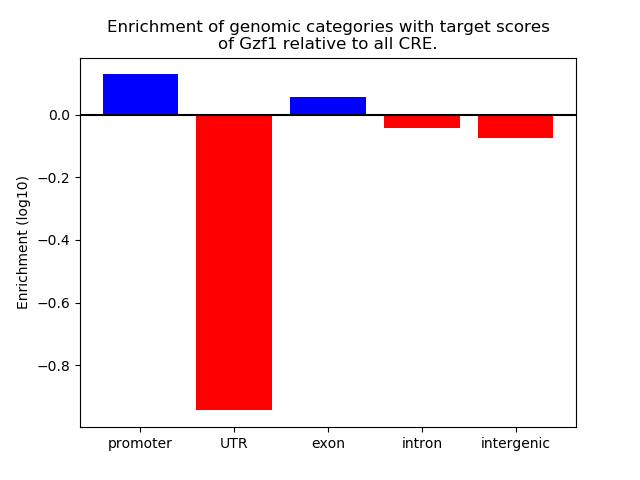
Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
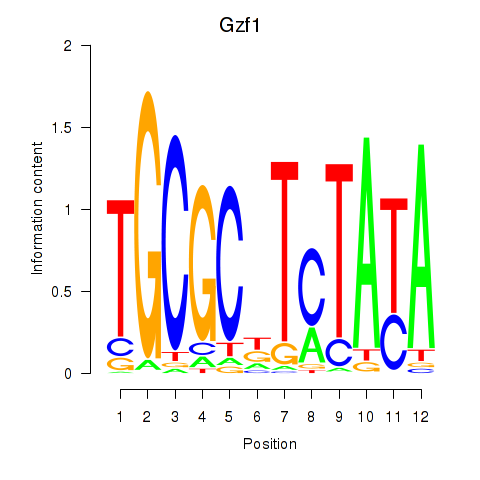
Results for Gzf1
Z-value: 1.27
Transcription factors associated with Gzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Gzf1
|
ENSMUSG00000027439.9 | GDNF-inducible zinc finger protein 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_148685781_148685971 | Gzf1 | 1264 | 0.390753 | -0.65 | 8.4e-08 | Click! |
| chr2_148685983_148686204 | Gzf1 | 1481 | 0.339685 | -0.62 | 3.5e-07 | Click! |
| chr2_148677887_148678489 | Gzf1 | 2835 | 0.209566 | 0.62 | 5.3e-07 | Click! |
| chr2_148677656_148677863 | Gzf1 | 3264 | 0.194164 | 0.52 | 5.4e-05 | Click! |
| chr2_148679740_148679915 | Gzf1 | 1196 | 0.412048 | 0.45 | 6.5e-04 | Click! |
Activity of the Gzf1 motif across conditions
Conditions sorted by the z-value of the Gzf1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
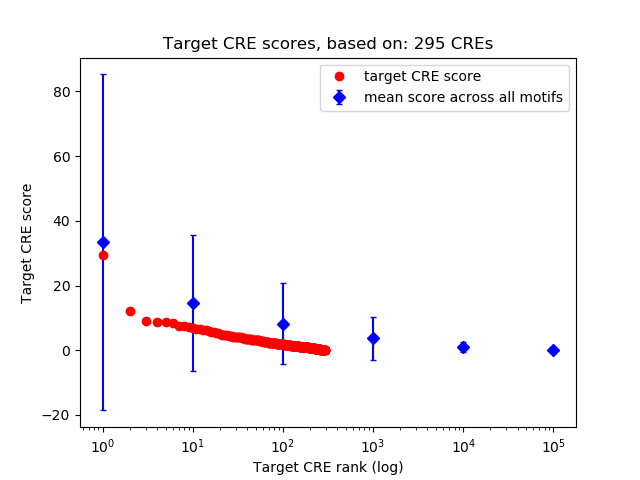
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_13590938_13591623 | 29.30 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
22416 |
0.14 |
| chr12_70345856_70346098 | 12.27 |
Trim9 |
tripartite motif-containing 9 |
1112 |
0.41 |
| chr5_39733465_39733625 | 9.09 |
Hs3st1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
21930 |
0.23 |
| chr16_72027511_72027683 | 8.73 |
Gm49667 |
predicted gene, 49667 |
148553 |
0.04 |
| chr12_113186726_113187124 | 8.62 |
Tmem121 |
transmembrane protein 121 |
995 |
0.4 |
| chr17_13654565_13655321 | 8.30 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
13948 |
0.15 |
| chr17_13589870_13590228 | 7.41 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
21185 |
0.15 |
| chr18_76469379_76469553 | 7.39 |
Gm50360 |
predicted gene, 50360 |
147642 |
0.04 |
| chr2_62807204_62807376 | 7.25 |
Gm13569 |
predicted gene 13569 |
1897 |
0.37 |
| chr17_13586214_13586541 | 6.98 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
17513 |
0.15 |
| chr10_59958140_59958359 | 6.61 |
Ddit4 |
DNA-damage-inducible transcript 4 |
6415 |
0.18 |
| chr17_13590288_13590503 | 6.55 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
21531 |
0.15 |
| chr2_170772344_170772652 | 6.39 |
Dok5 |
docking protein 5 |
40691 |
0.19 |
| chr11_68947505_68947921 | 6.38 |
Arhgef15 |
Rho guanine nucleotide exchange factor (GEF) 15 |
48 |
0.94 |
| chr15_41162973_41163124 | 5.80 |
4930555K19Rik |
RIKEN cDNA 4930555K19 gene |
10439 |
0.28 |
| chr13_71964060_71964241 | 5.60 |
Irx1 |
Iroquois homeobox 1 |
427 |
0.88 |
| chr14_80216319_80216498 | 5.49 |
Gm17923 |
predicted gene, 17923 |
1478 |
0.55 |
| chr2_102450292_102450505 | 5.39 |
Fjx1 |
four jointed box 1 |
2101 |
0.38 |
| chr6_112287813_112287964 | 5.36 |
5031434C07Rik |
RIKEN cDNA 5031434C07 gene |
948 |
0.51 |
| chr17_31057840_31058322 | 4.93 |
Abcg1 |
ATP binding cassette subfamily G member 1 |
307 |
0.82 |
| chr12_95694111_95694481 | 4.80 |
Flrt2 |
fibronectin leucine rich transmembrane protein 2 |
1061 |
0.51 |
| chr6_6884897_6885228 | 4.71 |
Dlx5 |
distal-less homeobox 5 |
2977 |
0.17 |
| chr7_144133020_144133171 | 4.56 |
Gm44999 |
predicted gene 44999 |
22010 |
0.2 |
| chr7_130367520_130367715 | 4.55 |
Gm5903 |
predicted gene 5903 |
13255 |
0.22 |
| chr5_99037530_99037747 | 4.41 |
Prkg2 |
protein kinase, cGMP-dependent, type II |
287 |
0.93 |
| chr11_77078150_77078604 | 4.40 |
Nsrp1 |
nuclear speckle regulatory protein 1 |
58 |
0.69 |
| chr4_98223390_98223614 | 4.33 |
Gm12692 |
predicted gene 12692 |
11788 |
0.21 |
| chr13_36613172_36613399 | 4.17 |
Gm46409 |
predicted gene, 46409 |
313 |
0.87 |
| chr2_100865612_100865778 | 4.15 |
Gm13810 |
predicted gene 13810 |
237652 |
0.02 |
| chr1_180725622_180726589 | 4.10 |
Acbd3 |
acyl-Coenzyme A binding domain containing 3 |
62 |
0.95 |
| chr12_85598845_85599442 | 4.09 |
Gm47863 |
predicted gene, 47863 |
48 |
0.58 |
| chr4_124861815_124862142 | 4.02 |
Maneal |
mannosidase, endo-alpha-like |
193 |
0.89 |
| chr7_6343440_6343815 | 4.00 |
Gm3854 |
predicted gene 3854 |
132 |
0.75 |
| chr7_4842196_4842588 | 3.96 |
Shisa7 |
shisa family member 7 |
2304 |
0.13 |
| chr1_173366946_173367141 | 3.76 |
Cadm3 |
cell adhesion molecule 3 |
590 |
0.7 |
| chr11_68341740_68341891 | 3.67 |
Ntn1 |
netrin 1 |
45011 |
0.14 |
| chr14_58071780_58071984 | 3.65 |
Fgf9 |
fibroblast growth factor 9 |
804 |
0.62 |
| chr1_47278444_47278595 | 3.60 |
Gm4852 |
predicted pseudogene 4852 |
87853 |
0.09 |
| chr12_65259311_65259487 | 3.52 |
Gm47973 |
predicted gene, 47973 |
7689 |
0.19 |
| chr2_57614579_57615021 | 3.44 |
Gm13532 |
predicted gene 13532 |
14428 |
0.2 |
| chr6_109874355_109874582 | 3.43 |
Gm44162 |
predicted gene, 44162 |
158921 |
0.04 |
| chr4_109763223_109763401 | 3.36 |
Gm12808 |
predicted gene 12808 |
8495 |
0.24 |
| chr7_63444022_63445137 | 3.34 |
4930554H23Rik |
RIKEN cDNA 4930554H23 gene |
50 |
0.67 |
| chr18_56015322_56015473 | 3.32 |
Gm6944 |
predicted gene 6944 |
43111 |
0.15 |
| chr11_100333236_100333409 | 3.19 |
Gast |
gastrin |
1085 |
0.28 |
| chr19_48204971_48205291 | 3.18 |
Sorcs3 |
sortilin-related VPS10 domain containing receptor 3 |
894 |
0.67 |
| chr18_44754207_44754358 | 3.14 |
Mcc |
mutated in colorectal cancers |
29624 |
0.17 |
| chr3_50381590_50381769 | 3.11 |
Slc7a11 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
10880 |
0.23 |
| chrX_7610162_7610425 | 3.10 |
Cacna1f |
calcium channel, voltage-dependent, alpha 1F subunit |
3149 |
0.1 |
| chr12_50809018_50809169 | 3.10 |
Gm48779 |
predicted gene, 48779 |
55996 |
0.15 |
| chr15_85678936_85679636 | 3.04 |
Lncppara |
long noncoding RNA near Ppara |
24487 |
0.12 |
| chr1_66387135_66387333 | 3.03 |
Map2 |
microtubule-associated protein 2 |
223 |
0.94 |
| chr5_63649551_63649743 | 3.02 |
Nwd2 |
NACHT and WD repeat domain containing 2 |
314 |
0.72 |
| chr7_73740150_73741122 | 3.01 |
A830073O21Rik |
RIKEN cDNA A830073O21 gene |
281 |
0.52 |
| chr3_56962365_56962516 | 2.94 |
Gm22269 |
predicted gene, 22269 |
91710 |
0.09 |
| chr11_20878789_20878940 | 2.89 |
Gm22807 |
predicted gene, 22807 |
30983 |
0.13 |
| chr11_94281227_94281543 | 2.88 |
Gm21885 |
predicted gene, 21885 |
488 |
0.76 |
| chr4_130055268_130055419 | 2.87 |
Col16a1 |
collagen, type XVI, alpha 1 |
288 |
0.87 |
| chr17_13587362_13587709 | 2.82 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
18671 |
0.15 |
| chr5_39417055_39417230 | 2.72 |
Gm5291 |
predicted gene 5291 |
14897 |
0.17 |
| chr4_31049383_31049534 | 2.68 |
Gm25705 |
predicted gene, 25705 |
125554 |
0.06 |
| chr5_139129572_139129764 | 2.68 |
Prkar1b |
protein kinase, cAMP dependent regulatory, type I beta |
91 |
0.96 |
| chr16_20619916_20620680 | 2.57 |
Camk2n2 |
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
989 |
0.25 |
| chr16_28444957_28445377 | 2.53 |
Fgf12 |
fibroblast growth factor 12 |
908 |
0.74 |
| chr17_67764573_67764743 | 2.45 |
Lama1 |
laminin, alpha 1 |
47748 |
0.16 |
| chr8_23411129_23412450 | 2.45 |
Sfrp1 |
secreted frizzled-related protein 1 |
287 |
0.94 |
| chr11_57694554_57694848 | 2.44 |
4933426K07Rik |
RIKEN cDNA 4933426K07 gene |
36875 |
0.11 |
| chr1_118954540_118954691 | 2.39 |
Mir6346 |
microRNA 6346 |
395 |
0.88 |
| chr12_32926365_32926853 | 2.39 |
Gm19056 |
predicted gene, 19056 |
3338 |
0.15 |
| chr7_83477873_83478111 | 2.38 |
Gm44991 |
predicted gene 44991 |
6284 |
0.21 |
| chr6_54008819_54008970 | 2.36 |
4921529L05Rik |
RIKEN cDNA 4921529L05 gene |
818 |
0.64 |
| chr15_95702329_95702480 | 2.36 |
Gm8843 |
predicted gene 8843 |
12789 |
0.16 |
| chr11_35914912_35915063 | 2.35 |
Wwc1 |
WW, C2 and coiled-coil domain containing 1 |
39275 |
0.16 |
| chr6_85451222_85452476 | 2.33 |
Pradc1 |
protease-associated domain containing 1 |
96 |
0.6 |
| chr7_84483886_84484050 | 2.33 |
Gm45175 |
predicted gene 45175 |
39417 |
0.11 |
| chr2_146810745_146810896 | 2.32 |
Gm14114 |
predicted gene 14114 |
28912 |
0.18 |
| chr11_47343450_47343810 | 2.31 |
Sgcd |
sarcoglycan, delta (dystrophin-associated glycoprotein) |
35892 |
0.23 |
| chr4_154960842_154960993 | 2.30 |
Hes5 |
hes family bHLH transcription factor 5 |
6 |
0.93 |
| chr4_63528950_63529291 | 2.26 |
Atp6v1g1 |
ATPase, H+ transporting, lysosomal V1 subunit G1 |
15652 |
0.12 |
| chr3_137762653_137762804 | 2.24 |
Mir6380 |
microRNA 6380 |
20887 |
0.13 |
| chr2_25290168_25290373 | 2.14 |
Lrrc26 |
leucine rich repeat containing 26 |
355 |
0.57 |
| chr9_14751478_14752576 | 2.12 |
Fut4 |
fucosyltransferase 4 |
366 |
0.74 |
| chr5_111218146_111218303 | 2.11 |
Ttc28 |
tetratricopeptide repeat domain 28 |
7234 |
0.2 |
| chr2_159002609_159002760 | 2.08 |
Gm44319 |
predicted gene, 44319 |
67179 |
0.12 |
| chr2_136108035_136108369 | 2.06 |
Gm14218 |
predicted gene 14218 |
28804 |
0.19 |
| chr9_73681333_73681484 | 2.05 |
Gm27148 |
predicted gene 27148 |
4716 |
0.31 |
| chr14_124192867_124193151 | 2.03 |
Fgf14 |
fibroblast growth factor 14 |
107 |
0.98 |
| chr13_81596951_81597255 | 1.96 |
Adgrv1 |
adhesion G protein-coupled receptor V1 |
12247 |
0.22 |
| chr10_79681206_79682337 | 1.95 |
Cdc34 |
cell division cycle 34 |
424 |
0.63 |
| chr10_98007193_98007368 | 1.92 |
Gm10754 |
predicted gene 10754 |
40190 |
0.21 |
| chr11_35927335_35927561 | 1.90 |
Wwc1 |
WW, C2 and coiled-coil domain containing 1 |
51736 |
0.13 |
| chr5_39772042_39772399 | 1.90 |
Hs3st1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
16745 |
0.22 |
| chr12_72069940_72071249 | 1.89 |
Gpr135 |
G protein-coupled receptor 135 |
531 |
0.72 |
| chr16_85086037_85086188 | 1.87 |
Gm49227 |
predicted gene, 49227 |
6001 |
0.22 |
| chr3_45377659_45378144 | 1.86 |
2610316D01Rik |
RIKEN cDNA 2610316D01 gene |
304 |
0.65 |
| chr18_12827295_12827569 | 1.85 |
Osbpl1a |
oxysterol binding protein-like 1A |
7525 |
0.19 |
| chr3_116512768_116512919 | 1.85 |
Dbt |
dihydrolipoamide branched chain transacylase E2 |
227 |
0.87 |
| chr7_16869301_16869754 | 1.84 |
Prkd2 |
protein kinase D2 |
275 |
0.8 |
| chr8_16965821_16965972 | 1.83 |
3110080E11Rik |
RIKEN cDNA 3110080E11 gene |
49714 |
0.18 |
| chr7_4844524_4844964 | 1.82 |
Shisa7 |
shisa family member 7 |
48 |
0.94 |
| chr3_93443881_93444908 | 1.82 |
Tchh |
trichohyalin |
2064 |
0.16 |
| chr4_99295280_99295454 | 1.82 |
Gm12689 |
predicted gene 12689 |
533 |
0.78 |
| chr8_4350654_4350950 | 1.76 |
Ccl25 |
chemokine (C-C motif) ligand 25 |
1214 |
0.32 |
| chr2_152142100_152142333 | 1.75 |
Tcf15 |
transcription factor 15 |
1345 |
0.37 |
| chr8_70487251_70487452 | 1.73 |
Tmem59l |
transmembrane protein 59-like |
7 |
0.92 |
| chr16_25407448_25407917 | 1.72 |
Gm18896 |
predicted gene, 18896 |
35591 |
0.22 |
| chr4_125583445_125583596 | 1.69 |
Mir692-2 |
microRNA 692-2 |
78771 |
0.09 |
| chr10_86492372_86492584 | 1.69 |
Syn3 |
synapsin III |
581 |
0.65 |
| chr4_6717891_6718227 | 1.67 |
Tox |
thymocyte selection-associated high mobility group box |
28708 |
0.19 |
| chr1_132283002_132283375 | 1.66 |
Gm7241 |
predicted pseudogene 7241 |
1265 |
0.3 |
| chr6_58907455_58907637 | 1.65 |
Herc3 |
hect domain and RLD 3 |
284 |
0.65 |
| chr6_29396003_29396644 | 1.63 |
Ccdc136 |
coiled-coil domain containing 136 |
14 |
0.79 |
| chr13_31792557_31792841 | 1.60 |
Gm11379 |
predicted gene 11379 |
4840 |
0.19 |
| chr14_18893044_18894412 | 1.58 |
Ube2e2 |
ubiquitin-conjugating enzyme E2E 2 |
10 |
0.98 |
| chr18_60926924_60927473 | 1.56 |
Camk2a |
calcium/calmodulin-dependent protein kinase II alpha |
249 |
0.89 |
| chr4_118127727_118128030 | 1.51 |
St3gal3 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
7004 |
0.17 |
| chr1_19951381_19951572 | 1.47 |
Gm37315 |
predicted gene, 37315 |
114072 |
0.07 |
| chr4_85831629_85831812 | 1.47 |
Gm25811 |
predicted gene, 25811 |
211774 |
0.02 |
| chr16_69469408_69469596 | 1.47 |
4930428D20Rik |
RIKEN cDNA 4930428D20 gene |
330879 |
0.01 |
| chr3_146345584_146345902 | 1.47 |
Gm43334 |
predicted gene 43334 |
21323 |
0.14 |
| chr15_9061253_9061404 | 1.47 |
Nadk2 |
NAD kinase 2, mitochondrial |
9932 |
0.22 |
| chr12_46075749_46076236 | 1.46 |
Gm24948 |
predicted gene, 24948 |
91649 |
0.09 |
| chr6_14900485_14900732 | 1.44 |
Foxp2 |
forkhead box P2 |
741 |
0.8 |
| chr16_52303296_52303447 | 1.44 |
Alcam |
activated leukocyte cell adhesion molecule |
6447 |
0.33 |
| chr15_74518307_74518458 | 1.44 |
Adgrb1 |
adhesion G protein-coupled receptor B1 |
1551 |
0.4 |
| chr1_152399789_152399970 | 1.43 |
Colgalt2 |
collagen beta(1-O)galactosyltransferase 2 |
49 |
0.98 |
| chr17_80372913_80373671 | 1.39 |
Gm10190 |
predicted gene 10190 |
249 |
0.62 |
| chr9_26454070_26454221 | 1.39 |
Gm48373 |
predicted gene, 48373 |
67843 |
0.11 |
| chr2_131040943_131041967 | 1.38 |
Gfra4 |
glial cell line derived neurotrophic factor family receptor alpha 4 |
2 |
0.94 |
| chr6_32328132_32328283 | 1.38 |
Plxna4os3 |
plexin A4, opposite strand 3 |
24294 |
0.24 |
| chr8_105268194_105269524 | 1.37 |
Hsf4 |
heat shock transcription factor 4 |
942 |
0.26 |
| chr16_25465879_25466059 | 1.36 |
Gm18896 |
predicted gene, 18896 |
22696 |
0.27 |
| chr5_137583806_137584189 | 1.35 |
Tfr2 |
transferrin receptor 2 |
712 |
0.39 |
| chr7_67071811_67071967 | 1.34 |
Gm17909 |
predicted gene, 17909 |
10954 |
0.19 |
| chr12_9166625_9166808 | 1.34 |
Ttc32 |
tetratricopeptide repeat domain 32 |
136688 |
0.04 |
| chr3_55756315_55756785 | 1.30 |
Mab21l1 |
mab-21-like 1 |
25960 |
0.18 |
| chr4_124036517_124036668 | 1.30 |
Gm12902 |
predicted gene 12902 |
110358 |
0.05 |
| chr7_129793883_129794034 | 1.29 |
Gm44778 |
predicted gene 44778 |
39088 |
0.2 |
| chr11_120223944_120224378 | 1.29 |
Gm11770 |
predicted gene 11770 |
7168 |
0.1 |
| chrX_140957056_140957443 | 1.26 |
Atg4a |
autophagy related 4A, cysteine peptidase |
299 |
0.68 |
| chr12_117159354_117159563 | 1.25 |
Gm10421 |
predicted gene 10421 |
7807 |
0.3 |
| chr13_35968213_35969019 | 1.24 |
Ppp1r3g |
protein phosphatase 1, regulatory subunit 3G |
670 |
0.57 |
| chr15_56692188_56692969 | 1.23 |
Has2os |
hyaluronan synthase 2, opposite strand |
974 |
0.55 |
| chr5_12498341_12498532 | 1.23 |
Sema3d |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
13481 |
0.21 |
| chr8_98365520_98365671 | 1.19 |
Gm7192 |
predicted gene 7192 |
140932 |
0.05 |
| chr1_170176895_170177131 | 1.17 |
Gm37502 |
predicted gene, 37502 |
759 |
0.47 |
| chr11_121702189_121702884 | 1.17 |
Metrnl |
meteorin, glial cell differentiation regulator-like |
125 |
0.97 |
| chr16_70535346_70535522 | 1.17 |
Gbe1 |
glucan (1,4-alpha-), branching enzyme 1 |
25560 |
0.24 |
| chr3_103733715_103734513 | 1.16 |
Olfml3 |
olfactomedin-like 3 |
115 |
0.93 |
| chr5_9100833_9101781 | 1.15 |
Tmem243 |
transmembrane protein 243, mitochondrial |
560 |
0.74 |
| chr17_36189653_36190417 | 1.15 |
H2-T3 |
histocompatibility 2, T region locus 3 |
87 |
0.86 |
| chr3_31308342_31308493 | 1.13 |
Slc7a14 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14 |
1961 |
0.31 |
| chr16_56920328_56920584 | 1.12 |
Lnp1 |
leukemia NUP98 fusion partner 1 |
7605 |
0.16 |
| chr10_85386973_85388092 | 1.12 |
Btbd11 |
BTB (POZ) domain containing 11 |
705 |
0.72 |
| chr19_8710179_8710404 | 1.11 |
Slc3a2 |
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
2694 |
0.07 |
| chr2_44565118_44565293 | 1.11 |
Gtdc1 |
glycosyltransferase-like domain containing 1 |
6142 |
0.33 |
| chr3_89225307_89226347 | 1.11 |
Thbs3 |
thrombospondin 3 |
423 |
0.5 |
| chr10_4006205_4006393 | 1.11 |
Gm16074 |
predicted gene 16074 |
13928 |
0.17 |
| chr4_45297019_45297341 | 1.10 |
Trmt10b |
tRNA methyltransferase 10B |
25 |
0.97 |
| chr10_3920376_3921003 | 1.09 |
Gm23023 |
predicted gene, 23023 |
14188 |
0.15 |
| chr17_76658153_76658304 | 1.09 |
Gm18366 |
predicted gene, 18366 |
163262 |
0.04 |
| chr7_46639145_46639386 | 1.08 |
Sergef |
secretion regulating guanine nucleotide exchange factor |
315 |
0.84 |
| chr14_118768381_118768663 | 1.06 |
Gm22379 |
predicted gene, 22379 |
6630 |
0.17 |
| chr2_131304736_131304905 | 1.05 |
Gm14233 |
predicted gene 14233 |
6275 |
0.12 |
| chr4_12287368_12287519 | 1.05 |
Gm11846 |
predicted gene 11846 |
16044 |
0.22 |
| chr18_73059570_73059810 | 1.04 |
Gm31908 |
predicted gene, 31908 |
138600 |
0.05 |
| chr6_71772911_71773062 | 1.04 |
Gm44769 |
predicted gene 44769 |
17682 |
0.12 |
| chr19_4508691_4508955 | 1.03 |
Pcx |
pyruvate carboxylase |
1649 |
0.27 |
| chr16_33739926_33740090 | 1.03 |
Heg1 |
heart development protein with EGF-like domains 1 |
8407 |
0.21 |
| chr11_59227641_59228869 | 1.02 |
Arf1 |
ADP-ribosylation factor 1 |
15 |
0.95 |
| chr8_14991358_14991509 | 1.00 |
Arhgef10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
1074 |
0.39 |
| chr5_70995664_70996254 | 0.99 |
Gabra2 |
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 2 |
17725 |
0.29 |
| chr6_38730546_38730697 | 0.99 |
Hipk2 |
homeodomain interacting protein kinase 2 |
12639 |
0.22 |
| chr6_97883597_97883757 | 0.96 |
Gm15531 |
predicted gene 15531 |
6862 |
0.26 |
| chr16_18588056_18588353 | 0.96 |
Tbx1 |
T-box 1 |
604 |
0.61 |
| chr5_24595821_24595972 | 0.96 |
Smarcd3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
953 |
0.34 |
| chr1_67065202_67065417 | 0.95 |
Lancl1 |
LanC (bacterial lantibiotic synthetase component C)-like 1 |
26437 |
0.18 |
| chr1_163314015_163314239 | 0.95 |
Prrx1 |
paired related homeobox 1 |
417 |
0.83 |
| chrX_48170424_48170619 | 0.95 |
Sash3 |
SAM and SH3 domain containing 3 |
24085 |
0.15 |
| chr4_128991868_128992156 | 0.94 |
Ak2 |
adenylate kinase 2 |
54 |
0.97 |
| chrX_103289004_103289290 | 0.93 |
Gm19200 |
predicted gene, 19200 |
177 |
0.86 |
| chr7_124704634_124704798 | 0.93 |
3100003L05Rik |
RIKEN cDNA 3100003L05 gene |
4219 |
0.35 |
| chr5_142509557_142509714 | 0.91 |
Radil |
Ras association and DIL domains |
24 |
0.97 |
| chr18_12917222_12917373 | 0.91 |
Gm5687 |
predicted gene 5687 |
18870 |
0.16 |
| chr5_105655269_105655420 | 0.91 |
Lrrc8d |
leucine rich repeat containing 8D |
44625 |
0.14 |
| chr10_67536867_67537245 | 0.89 |
Egr2 |
early growth response 2 |
813 |
0.53 |
| chr11_101785577_101785749 | 0.87 |
Etv4 |
ets variant 4 |
292 |
0.88 |
| chr15_61448570_61448721 | 0.87 |
Gm24696 |
predicted gene, 24696 |
90641 |
0.09 |
| chr1_47604096_47604257 | 0.87 |
Gm37196 |
predicted gene, 37196 |
1820 |
0.44 |
| chr4_70058924_70059247 | 0.87 |
Gm11225 |
predicted gene 11225 |
11928 |
0.28 |
| chrX_94990647_94990838 | 0.86 |
Gm371 |
predicted pseudogene 371 |
7941 |
0.22 |
| chr7_98692838_98693482 | 0.86 |
Gm15506 |
predicted gene 15506 |
9274 |
0.13 |
| chr4_129959595_129960137 | 0.83 |
1700003M07Rik |
RIKEN cDNA 1700003M07 gene |
508 |
0.71 |
| chr5_139907692_139908613 | 0.83 |
Elfn1 |
leucine rich repeat and fibronectin type III, extracellular 1 |
209 |
0.93 |
| chr1_74092335_74092587 | 0.81 |
Tns1 |
tensin 1 |
1110 |
0.49 |
| chr3_7424444_7424644 | 0.79 |
Pkia |
protein kinase inhibitor, alpha |
4514 |
0.21 |
| chr2_4442210_4442396 | 0.79 |
Gm13175 |
predicted gene 13175 |
24367 |
0.17 |
| chr8_16114909_16115569 | 0.78 |
Mir3106 |
microRNA 3106 |
53599 |
0.16 |
| chr16_34572272_34572475 | 0.78 |
Kalrn |
kalirin, RhoGEF kinase |
1159 |
0.62 |
| chr14_22760839_22761006 | 0.77 |
Gm7473 |
predicted gene 7473 |
14322 |
0.29 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.3 | 4.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 1.1 | 2.1 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 1.0 | 5.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.8 | 2.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.8 | 2.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.7 | 2.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.5 | 1.6 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.4 | 2.5 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.3 | 2.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 1.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 0.9 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.3 | 1.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.3 | 0.8 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.3 | 0.8 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.3 | 0.8 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 1.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.3 | 2.6 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.2 | 0.9 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 2.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.2 | 0.2 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.2 | 1.4 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.2 | 1.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 0.6 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 3.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.2 | 0.5 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 | 0.5 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.1 | 2.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 1.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.8 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.3 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 | 0.5 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 2.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.4 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 1.5 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.1 | 0.3 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.4 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 1.2 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.1 | 1.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.9 | GO:1904869 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.3 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 1.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 0.8 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.3 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 2.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.1 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.4 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 3.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.2 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 1.7 | GO:2001258 | negative regulation of cation channel activity(GO:2001258) |
| 0.1 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.2 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.5 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 | 0.2 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.2 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 | 0.6 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.3 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 2.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 1.0 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.3 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.0 | 0.1 | GO:0033505 | floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 3.2 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.3 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.4 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:1903147 | negative regulation of macromitophagy(GO:1901525) negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.1 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.0 | 0.0 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.3 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.1 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.3 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.0 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.7 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.0 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.0 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.8 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.1 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.1 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.0 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.0 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0030214 | hyaluronan catabolic process(GO:0030214) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.4 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) regulation of epinephrine secretion(GO:0014060) negative regulation of epinephrine secretion(GO:0032811) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.3 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.6 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.1 | 0.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 2.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 2.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 6.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.9 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 2.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 9.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 2.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 5.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 1.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 2.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 1.0 | 2.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.5 | 1.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.5 | 2.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.4 | 2.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.3 | 4.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.3 | 8.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 0.9 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.3 | 0.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 3.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 0.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 0.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 2.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 7.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 1.9 | GO:0052859 | dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.1 | 2.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.3 | GO:0018594 | mono-butyltin dioxygenase activity(GO:0018586) tri-n-butyltin dioxygenase activity(GO:0018588) di-n-butyltin dioxygenase activity(GO:0018589) methylsilanetriol hydroxylase activity(GO:0018590) methyl tertiary butyl ether 3-monooxygenase activity(GO:0018591) 4-nitrocatechol 4-monooxygenase activity(GO:0018592) 4-chlorophenoxyacetate monooxygenase activity(GO:0018593) tert-butanol 2-monooxygenase activity(GO:0018594) alpha-pinene monooxygenase activity(GO:0018595) dimethylsilanediol hydroxylase activity(GO:0018596) ammonia monooxygenase activity(GO:0018597) hydroxymethylsilanetriol oxidase activity(GO:0018598) 2-hydroxyisobutyrate 3-monooxygenase activity(GO:0018599) alpha-pinene dehydrogenase activity(GO:0018600) bisphenol A hydroxylase B activity(GO:0034559) 2,2-bis(4-hydroxyphenyl)-1-propanol hydroxylase activity(GO:0034562) 9-fluorenone-3,4-dioxygenase activity(GO:0034786) anthracene 9,10-dioxygenase activity(GO:0034816) 2-(methylthio)benzothiazole monooxygenase activity(GO:0034857) 2-hydroxybenzothiazole monooxygenase activity(GO:0034858) benzothiazole monooxygenase activity(GO:0034859) 2,6-dihydroxybenzothiazole monooxygenase activity(GO:0034862) pinacolone 5-monooxygenase activity(GO:0034870) thioacetamide S-oxygenase activity(GO:0034873) thioacetamide S-oxide S-oxygenase activity(GO:0034874) endosulfan monooxygenase I activity(GO:0034888) N-nitrodimethylamine hydroxylase activity(GO:0034893) 4-(1-ethyl-1,4-dimethyl-pentyl)phenol monoxygenase activity(GO:0034897) endosulfan ether monooxygenase activity(GO:0034903) pyrene 4,5-monooxygenase activity(GO:0034925) pyrene 1,2-monooxygenase activity(GO:0034927) 1-hydroxypyrene 6,7-monooxygenase activity(GO:0034928) 1-hydroxypyrene 7,8-monooxygenase activity(GO:0034929) phenylboronic acid monooxygenase activity(GO:0034950) spheroidene monooxygenase activity(GO:0043823) |
| 0.1 | 1.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.2 | GO:0035276 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) ethanol binding(GO:0035276) |
| 0.1 | 0.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.2 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 5.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 4.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 2.8 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.5 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.1 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 3.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 2.7 | GO:0003729 | mRNA binding(GO:0003729) |
| 0.0 | 0.1 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 1.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 2.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 3.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 6.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 4.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 2.5 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.2 | 2.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 0.4 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 2.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 6.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 1.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |