Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
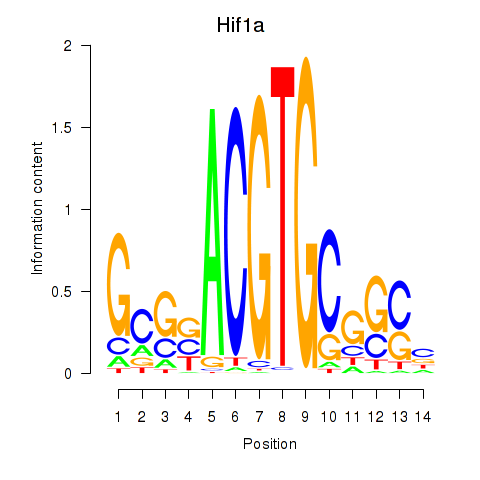
Results for Hif1a
Z-value: 0.64
Transcription factors associated with Hif1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hif1a
|
ENSMUSG00000021109.7 | hypoxia inducible factor 1, alpha subunit |
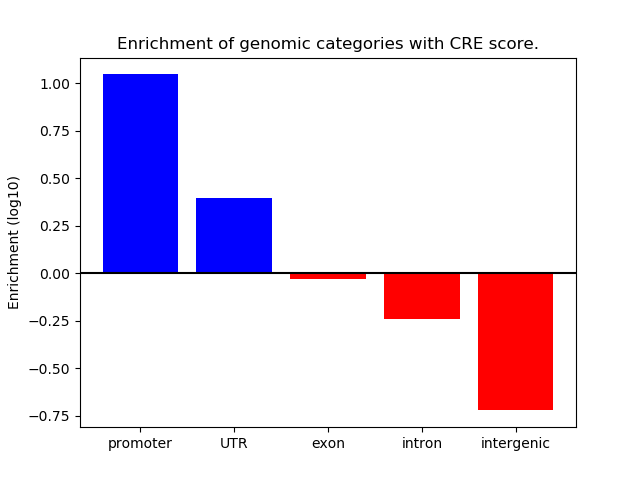
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_73907701_73907880 | Hif1a | 114 | 0.960521 | -0.65 | 8.4e-08 | Click! |
| chr12_73907449_73907656 | Hif1a | 352 | 0.858120 | -0.64 | 1.7e-07 | Click! |
| chr12_73907945_73908301 | Hif1a | 219 | 0.925032 | -0.60 | 1.4e-06 | Click! |
| chr12_73908374_73908672 | Hif1a | 619 | 0.696810 | -0.44 | 8.0e-04 | Click! |
| chr12_73905175_73905337 | Hif1a | 2648 | 0.239489 | -0.02 | 8.8e-01 | Click! |
Activity of the Hif1a motif across conditions
Conditions sorted by the z-value of the Hif1a motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
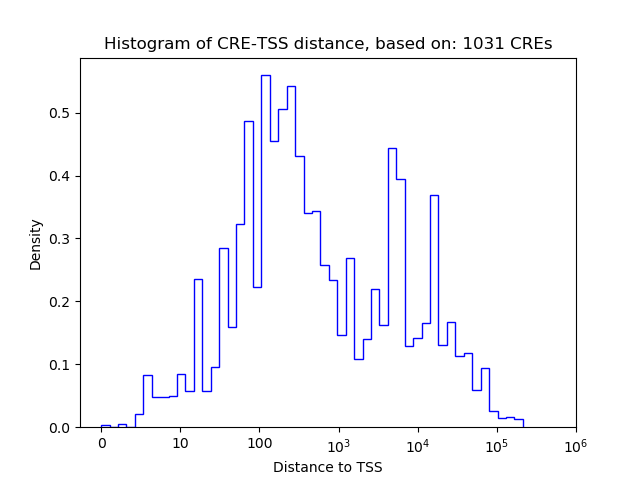
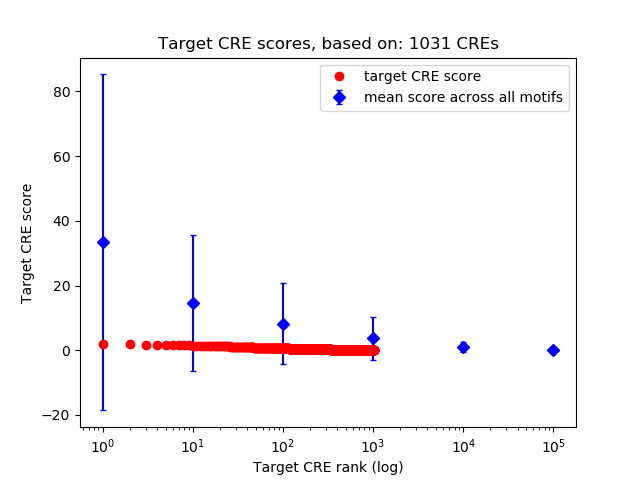
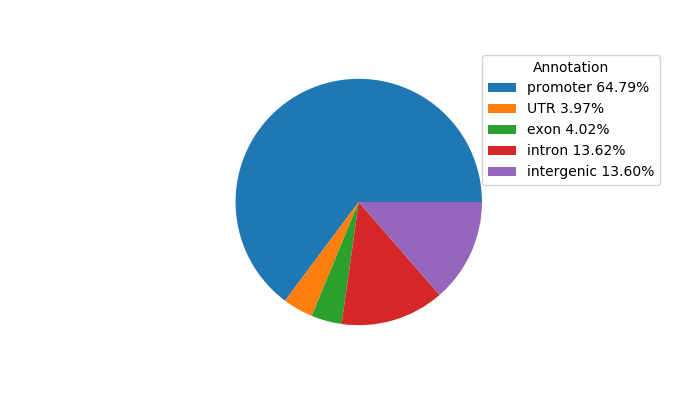
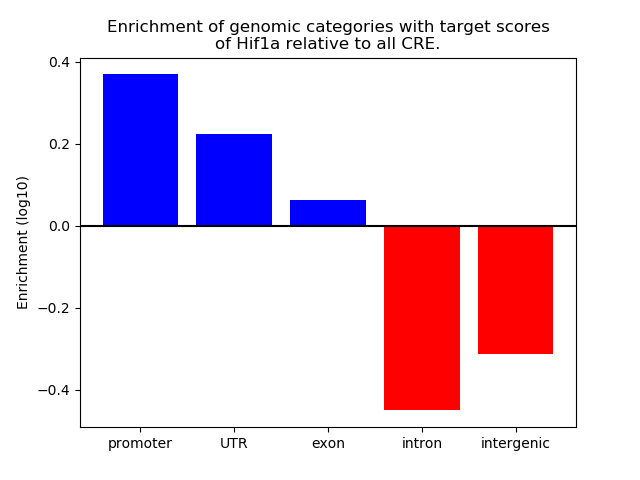
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr14_6756349_6757421 | 1.93 |
Gm3636 |
predicted gene 3636 |
14553 |
0.14 |
| chr14_4018192_4019443 | 1.80 |
Gm5796 |
predicted gene 5796 |
5124 |
0.14 |
| chr14_3332417_3333262 | 1.71 |
Gm2956 |
predicted gene 2956 |
212 |
0.91 |
| chr14_4334095_4335404 | 1.64 |
2610042L04Rik |
RIKEN cDNA 2610042L04 gene |
14 |
0.97 |
| chr14_3412209_3412992 | 1.61 |
Gm10409 |
predicted gene 10409 |
14 |
0.96 |
| chr14_4726126_4727100 | 1.58 |
Gm3252 |
predicted gene 3252 |
162 |
0.93 |
| chr14_6108337_6108836 | 1.54 |
Gm3468 |
predicted gene 3468 |
79 |
0.95 |
| chr14_3809454_3810456 | 1.52 |
Gm3002 |
predicted gene 3002 |
119 |
0.95 |
| chr11_102819100_102819714 | 1.50 |
Gjc1 |
gap junction protein, gamma 1 |
197 |
0.89 |
| chr14_3571400_3572424 | 1.45 |
Gm3005 |
predicted gene 3005 |
111 |
0.96 |
| chr14_7105202_7106057 | 1.43 |
Gm3696 |
predicted gene 3696 |
5008 |
0.15 |
| chr1_179667564_179668717 | 1.42 |
Sccpdh |
saccharopine dehydrogenase (putative) |
70 |
0.98 |
| chr7_4521231_4521527 | 1.38 |
Tnni3 |
troponin I, cardiac 3 |
812 |
0.37 |
| chr14_7488010_7488824 | 1.34 |
Gm3752 |
predicted gene 3752 |
4655 |
0.16 |
| chr14_4181957_4182899 | 1.34 |
Gm2974 |
predicted gene 2974 |
148 |
0.94 |
| chr4_107830355_107830587 | 1.33 |
Lrp8os1 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor, opposite strand 1 |
373 |
0.53 |
| chr18_24714654_24714883 | 1.29 |
Fhod3 |
formin homology 2 domain containing 3 |
5323 |
0.18 |
| chr14_7658268_7659211 | 1.29 |
Gm10128 |
predicted gene 10128 |
14767 |
0.16 |
| chr14_3651827_3652389 | 1.26 |
Gm3020 |
predicted gene 3020 |
78 |
0.96 |
| chr14_19584518_19585206 | 1.24 |
Gm2237 |
predicted gene 2237 |
273 |
0.84 |
| chr14_6426499_6427300 | 1.23 |
Lamtor3-ps |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3, pseudogene |
14417 |
0.14 |
| chr8_111094195_111095226 | 1.20 |
Pdpr |
pyruvate dehydrogenase phosphatase regulatory subunit |
55 |
0.95 |
| chr14_4498574_4499972 | 1.20 |
Gm3173 |
predicted gene 3173 |
15485 |
0.11 |
| chr4_139637979_139638130 | 1.17 |
Mir7020 |
microRNA 7020 |
5988 |
0.15 |
| chr8_15027100_15028701 | 1.16 |
Gm37844 |
predicted gene, 37844 |
522 |
0.44 |
| chr9_87015281_87015918 | 1.13 |
Ripply2 |
ripply transcriptional repressor 2 |
37 |
0.97 |
| chr14_5501458_5502048 | 1.12 |
Gm3488 |
predicted gene, 3488 |
79 |
0.95 |
| chr7_120102194_120103051 | 1.11 |
Tmem159 |
transmembrane protein 159 |
132 |
0.86 |
| chr11_47988672_47989535 | 1.09 |
Sgcd |
sarcoglycan, delta (dystrophin-associated glycoprotein) |
134 |
0.98 |
| chr8_111144981_111146043 | 1.08 |
9430091E24Rik |
RIKEN cDNA 9430091E24 gene |
32 |
0.97 |
| chr9_64110607_64110889 | 1.06 |
Lctl |
lactase-like |
6399 |
0.12 |
| chr7_25010372_25010757 | 1.06 |
Atp1a3 |
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
4606 |
0.12 |
| chr14_6287027_6287952 | 1.06 |
Gm3411 |
predicted gene 3411 |
239 |
0.89 |
| chr6_148443752_148444889 | 1.03 |
Tmtc1 |
transmembrane and tetratricopeptide repeat containing 1 |
69 |
0.98 |
| chr11_60104775_60106860 | 1.02 |
Rai1 |
retinoic acid induced 1 |
640 |
0.67 |
| chr8_29219544_29220019 | 1.00 |
Unc5d |
unc-5 netrin receptor D |
145 |
0.97 |
| chr5_74147781_74147946 | 0.99 |
A330058E17Rik |
RIKEN cDNA A330058E17 gene |
28187 |
0.11 |
| chr14_5148821_5149378 | 0.99 |
Gm3317 |
predicted gene 3317 |
15436 |
0.11 |
| chr2_144010017_144011636 | 0.97 |
Rrbp1 |
ribosome binding protein 1 |
437 |
0.82 |
| chr14_7027128_7027881 | 0.95 |
Gm10406 |
predicted gene 10406 |
55 |
0.96 |
| chr14_7174332_7174673 | 0.92 |
Gm3512 |
predicted gene 3512 |
31 |
0.97 |
| chr5_25498778_25500023 | 0.90 |
Kmt2c |
lysine (K)-specific methyltransferase 2C |
617 |
0.62 |
| chr1_22533152_22534195 | 0.87 |
Rims1 |
regulating synaptic membrane exocytosis 1 |
21144 |
0.28 |
| chr3_135417369_135417520 | 0.86 |
Cisd2 |
CDGSH iron sulfur domain 2 |
5635 |
0.12 |
| chrX_136666320_136667042 | 0.85 |
Tceal3 |
transcription elongation factor A (SII)-like 3 |
70 |
0.96 |
| chr7_45234372_45235480 | 0.84 |
Cd37 |
CD37 antigen |
532 |
0.49 |
| chr7_143107542_143107900 | 0.84 |
Kcnq1 |
potassium voltage-gated channel, subfamily Q, member 1 |
170 |
0.91 |
| chr2_61812475_61812626 | 0.82 |
Tbr1 |
T-box brain gene 1 |
6277 |
0.19 |
| chr14_6889264_6890534 | 0.81 |
Gm3667 |
predicted gene 3667 |
63 |
0.97 |
| chr14_50061711_50061923 | 0.81 |
Gm37194 |
predicted gene, 37194 |
457 |
0.76 |
| chr1_42698067_42698308 | 0.80 |
Pou3f3 |
POU domain, class 3, transcription factor 3 |
2419 |
0.19 |
| chr1_42952543_42953742 | 0.79 |
Gpr45 |
G protein-coupled receptor 45 |
3 |
0.98 |
| chr14_4855366_4855912 | 0.79 |
Gm3264 |
predicted gene 3264 |
63 |
0.96 |
| chr8_69881901_69882713 | 0.79 |
Cilp2 |
cartilage intermediate layer protein 2 |
5380 |
0.11 |
| chr9_114383017_114383329 | 0.79 |
Crtap |
cartilage associated protein |
4952 |
0.16 |
| chr13_18947816_18948472 | 0.79 |
Amph |
amphiphysin |
61 |
0.97 |
| chr3_110249839_110251122 | 0.78 |
C130013H08Rik |
RIKEN cDNA C130013H08 gene |
465 |
0.49 |
| chr3_73053654_73054054 | 0.78 |
Slitrk3 |
SLIT and NTRK-like family, member 3 |
3089 |
0.24 |
| chr5_37242202_37242734 | 0.77 |
Crmp1 |
collapsin response mediator protein 1 |
266 |
0.92 |
| chr2_13010261_13011265 | 0.76 |
Gm37811 |
predicted gene, 37811 |
877 |
0.35 |
| chr4_143211895_143213040 | 0.76 |
Prdm2 |
PR domain containing 2, with ZNF domain |
238 |
0.9 |
| chr12_33957808_33958154 | 0.75 |
Twist1 |
twist basic helix-loop-helix transcription factor 1 |
310 |
0.91 |
| chr13_116297452_116297610 | 0.75 |
Isl1 |
ISL1 transcription factor, LIM/homeodomain |
5820 |
0.25 |
| chr12_112781968_112782119 | 0.75 |
Ahnak2 |
AHNAK nucleoprotein 2 |
1032 |
0.36 |
| chr9_72924923_72926161 | 0.74 |
Pygo1 |
pygopus 1 |
103 |
0.93 |
| chr15_95702329_95702480 | 0.74 |
Gm8843 |
predicted gene 8843 |
12789 |
0.16 |
| chr13_48966851_48968700 | 0.74 |
Fam120a |
family with sequence similarity 120, member A |
242 |
0.95 |
| chr10_78747019_78747870 | 0.73 |
Gm30400 |
predicted gene, 30400 |
285 |
0.85 |
| chr1_85325328_85325596 | 0.72 |
Gm16025 |
predicted gene 16025 |
1267 |
0.31 |
| chr2_22895114_22896094 | 0.72 |
Pdss1 |
prenyl (solanesyl) diphosphate synthase, subunit 1 |
9 |
0.63 |
| chrX_110814525_110815208 | 0.71 |
Pou3f4 |
POU domain, class 3, transcription factor 4 |
586 |
0.77 |
| chr11_5788301_5789276 | 0.71 |
Dbnl |
drebrin-like |
258 |
0.86 |
| chr9_8003315_8004975 | 0.71 |
Yap1 |
yes-associated protein 1 |
245 |
0.79 |
| chr14_5460033_5460567 | 0.69 |
Gm3194 |
predicted gene 3194 |
4833 |
0.14 |
| chr9_100095718_100095869 | 0.68 |
4930519F24Rik |
RIKEN cDNA 4930519F24 gene |
72145 |
0.1 |
| chr9_90163003_90164118 | 0.68 |
Adamts7 |
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
334 |
0.85 |
| chr17_14279299_14279731 | 0.68 |
Smoc2 |
SPARC related modular calcium binding 2 |
9 |
0.98 |
| chr14_5812521_5813051 | 0.67 |
Gm3383 |
predicted gene 3383 |
4804 |
0.13 |
| chr4_101419063_101420491 | 0.66 |
Ak4 |
adenylate kinase 4 |
44 |
0.97 |
| chr8_84742133_84743295 | 0.66 |
G430095P16Rik |
RIKEN cDNA G430095P16 gene |
19707 |
0.09 |
| chr7_35554990_35556314 | 0.65 |
B230322F03Rik |
RIKEN cDNA B230322F03 gene |
285 |
0.62 |
| chr11_121259259_121260714 | 0.65 |
Foxk2 |
forkhead box K2 |
4 |
0.96 |
| chr5_142367069_142368113 | 0.65 |
Foxk1 |
forkhead box K1 |
33906 |
0.18 |
| chr17_43629678_43630114 | 0.64 |
Tdrd6 |
tudor domain containing 6 |
370 |
0.84 |
| chr15_88733665_88734619 | 0.64 |
Brd1 |
bromodomain containing 1 |
88 |
0.97 |
| chr7_18957517_18958673 | 0.63 |
Nanos2 |
nanos C2HC-type zinc finger 2 |
29305 |
0.06 |
| chr15_57745156_57745804 | 0.63 |
9330154K18Rik |
RIKEN cDNA 9330154K18 gene |
6814 |
0.22 |
| chr17_27555089_27556137 | 0.63 |
Hmga1 |
high mobility group AT-hook 1 |
884 |
0.27 |
| chr13_55471184_55472974 | 0.62 |
Mir6944 |
microRNA 6944 |
5697 |
0.08 |
| chr10_85102468_85103421 | 0.61 |
Tmem263 |
transmembrane protein 263 |
201 |
0.74 |
| chr16_91223532_91223683 | 0.61 |
Gm49614 |
predicted gene, 49614 |
681 |
0.54 |
| chr4_136637166_136638197 | 0.61 |
Gm37583 |
predicted gene, 37583 |
1901 |
0.22 |
| chr14_5070417_5071423 | 0.61 |
Gm8281 |
predicted gene, 8281 |
120 |
0.95 |
| chr3_80800657_80801686 | 0.60 |
Gria2 |
glutamate receptor, ionotropic, AMPA2 (alpha 2) |
1408 |
0.52 |
| chr5_71657445_71657639 | 0.60 |
Gabra4 |
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
278 |
0.73 |
| chr2_75246191_75246393 | 0.60 |
2600014E21Rik |
RIKEN cDNA 2600014E21 gene |
2108 |
0.3 |
| chr1_33907825_33908362 | 0.60 |
Dst |
dystonin |
132 |
0.68 |
| chr18_25504451_25505014 | 0.58 |
Celf4 |
CUGBP, Elav-like family member 4 |
3493 |
0.34 |
| chr13_99516425_99517155 | 0.58 |
Gm26559 |
predicted gene, 26559 |
161 |
0.62 |
| chr18_34006863_34007553 | 0.57 |
Epb41l4a |
erythrocyte membrane protein band 4.1 like 4a |
242 |
0.92 |
| chr8_35328966_35329165 | 0.56 |
Ppp1r3b |
protein phosphatase 1, regulatory subunit 3B |
46674 |
0.11 |
| chr14_65423658_65423809 | 0.56 |
Pnoc |
prepronociceptin |
1427 |
0.44 |
| chr16_34921371_34921807 | 0.56 |
Mylk |
myosin, light polypeptide kinase |
8753 |
0.19 |
| chr11_3331774_3332581 | 0.56 |
Pik3ip1 |
phosphoinositide-3-kinase interacting protein 1 |
68 |
0.93 |
| chr14_122490496_122491140 | 0.56 |
Gm10837 |
predicted gene 10837 |
232 |
0.87 |
| chr2_153258876_153259060 | 0.56 |
Pofut1 |
protein O-fucosyltransferase 1 |
1396 |
0.34 |
| chr3_57574902_57575246 | 0.56 |
Wwtr1 |
WW domain containing transcription regulator 1 |
754 |
0.61 |
| chr9_107400062_107400379 | 0.56 |
Cacna2d2 |
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
151 |
0.93 |
| chr10_80225959_80227288 | 0.55 |
Pwwp3a |
PWWP domain containing 3A, DNA repair factor |
32 |
0.94 |
| chr2_152143351_152143985 | 0.55 |
Tcf15 |
transcription factor 15 |
107 |
0.96 |
| chr12_117533503_117534508 | 0.55 |
Rapgef5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
117 |
0.97 |
| chr1_181352151_181352477 | 0.54 |
Cnih3 |
cornichon family AMPA receptor auxiliary protein 3 |
314 |
0.88 |
| chr11_7055619_7056056 | 0.54 |
Adcy1 |
adenylate cyclase 1 |
7652 |
0.26 |
| chr3_129204151_129204986 | 0.54 |
Pitx2 |
paired-like homeodomain transcription factor 2 |
621 |
0.7 |
| chr7_46179909_46180259 | 0.53 |
Abcc8 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
51 |
0.96 |
| chr11_51855495_51856955 | 0.53 |
Jade2 |
jade family PHD finger 2 |
900 |
0.57 |
| chr6_39256581_39257096 | 0.53 |
Gm26008 |
predicted gene, 26008 |
9104 |
0.16 |
| chr7_6171760_6172680 | 0.53 |
Zfp444 |
zinc finger protein 444 |
6 |
0.95 |
| chrX_157613038_157613189 | 0.52 |
Gm15173 |
predicted gene 15173 |
8 |
0.96 |
| chr18_65430370_65431071 | 0.52 |
Malt1 |
MALT1 paracaspase |
218 |
0.9 |
| chr5_136038932_136039111 | 0.52 |
Upk3b |
uroplakin 3B |
525 |
0.64 |
| chr13_71558486_71558841 | 0.52 |
Gm47811 |
predicted gene, 47811 |
66311 |
0.11 |
| chr10_24479227_24479575 | 0.52 |
Gm15271 |
predicted gene 15271 |
31911 |
0.15 |
| chr12_45563232_45563656 | 0.52 |
Gm48517 |
predicted gene, 48517 |
9843 |
0.27 |
| chr2_152093402_152093892 | 0.52 |
Srxn1 |
sulfiredoxin 1 homolog (S. cerevisiae) |
11869 |
0.13 |
| chr3_101109480_101110654 | 0.51 |
Ptgfrn |
prostaglandin F2 receptor negative regulator |
211 |
0.94 |
| chr1_59376261_59376435 | 0.51 |
Gm29016 |
predicted gene 29016 |
2582 |
0.29 |
| chr9_114528016_114528485 | 0.51 |
C130032M10Rik |
RIKEN cDNA C130032M10 gene |
12553 |
0.16 |
| chr7_5080037_5080839 | 0.50 |
Epn1 |
epsin 1 |
145 |
0.87 |
| chr2_37775771_37775969 | 0.50 |
Crb2 |
crumbs family member 2 |
379 |
0.88 |
| chr17_55445138_55445530 | 0.49 |
St6gal2 |
beta galactoside alpha 2,6 sialyltransferase 2 |
48 |
0.98 |
| chr12_108794058_108794786 | 0.49 |
Yy1 |
YY1 transcription factor |
1449 |
0.23 |
| chr17_28271837_28272180 | 0.49 |
Ppard |
peroxisome proliferator activator receptor delta |
111 |
0.94 |
| chr11_82388737_82388922 | 0.49 |
Tmem132e |
transmembrane protein 132E |
71 |
0.97 |
| chr2_156614281_156614628 | 0.48 |
Dlgap4 |
DLG associated protein 4 |
749 |
0.35 |
| chr7_138909400_138910240 | 0.48 |
Bnip3 |
BCL2/adenovirus E1B interacting protein 3 |
301 |
0.83 |
| chr6_80622673_80623041 | 0.48 |
Gm44231 |
predicted gene, 44231 |
32172 |
0.23 |
| chr6_48629216_48629950 | 0.48 |
AI854703 |
expressed sequence AI854703 |
1648 |
0.16 |
| chr12_20415451_20415611 | 0.47 |
Gm49929 |
predicted gene, 49929 |
406 |
0.77 |
| chr8_117157446_117158085 | 0.47 |
Gan |
giant axonal neuropathy |
370 |
0.87 |
| chr2_48538627_48539050 | 0.47 |
Gm13481 |
predicted gene 13481 |
81593 |
0.1 |
| chr18_35920048_35920506 | 0.46 |
Gm50405 |
predicted gene, 50405 |
3018 |
0.15 |
| chr14_122463926_122464344 | 0.46 |
Zic5 |
zinc finger protein of the cerebellum 5 |
1542 |
0.26 |
| chr4_154329867_154330297 | 0.46 |
Prdm16 |
PR domain containing 16 |
4770 |
0.18 |
| chr10_80398762_80400128 | 0.46 |
Mbd3 |
methyl-CpG binding domain protein 3 |
23 |
0.93 |
| chr13_73265452_73265676 | 0.46 |
Irx4 |
Iroquois homeobox 4 |
5067 |
0.17 |
| chr6_72898945_72900607 | 0.46 |
Kcmf1 |
potassium channel modulatory factor 1 |
73 |
0.97 |
| chr4_49844763_49845632 | 0.45 |
Grin3a |
glutamate receptor ionotropic, NMDA3A |
352 |
0.92 |
| chr18_40256699_40257075 | 0.45 |
Kctd16 |
potassium channel tetramerisation domain containing 16 |
75 |
0.96 |
| chr4_82513118_82513685 | 0.45 |
Gm11266 |
predicted gene 11266 |
5385 |
0.22 |
| chr2_147188117_147188295 | 0.45 |
6430503K07Rik |
RIKEN cDNA 6430503K07 gene |
782 |
0.52 |
| chr13_94868641_94869006 | 0.45 |
Otp |
orthopedia homeobox |
6779 |
0.21 |
| chr8_57354026_57354218 | 0.45 |
5033428I22Rik |
RIKEN cDNA 5033428I22 gene |
13322 |
0.13 |
| chr7_28294529_28294993 | 0.44 |
Dll3 |
delta like canonical Notch ligand 3 |
280 |
0.78 |
| chr7_49088814_49088965 | 0.44 |
Gm32849 |
predicted gene, 32849 |
3806 |
0.23 |
| chr2_79585158_79585309 | 0.44 |
Itprid2 |
ITPR interacting domain containing 2 |
50119 |
0.16 |
| chr12_102729973_102730175 | 0.43 |
Gm28373 |
predicted gene 28373 |
2741 |
0.12 |
| chr19_24031055_24031408 | 0.43 |
Fam189a2 |
family with sequence similarity 189, member A2 |
212 |
0.93 |
| chr9_80332183_80332515 | 0.42 |
Myo6 |
myosin VI |
25435 |
0.21 |
| chr6_99726441_99726592 | 0.42 |
Prok2 |
prokineticin 2 |
124 |
0.9 |
| chr9_107231249_107231435 | 0.42 |
Dock3 |
dedicator of cyto-kinesis 3 |
240 |
0.6 |
| chr5_110652819_110653593 | 0.42 |
Noc4l |
NOC4 like |
53 |
0.74 |
| chr14_69390979_69391250 | 0.42 |
Gm21451 |
predicted gene, 21451 |
260 |
0.86 |
| chr18_46109567_46109759 | 0.42 |
Gm38337 |
predicted gene, 38337 |
3596 |
0.27 |
| chr14_33319515_33320532 | 0.41 |
Arhgap22 |
Rho GTPase activating protein 22 |
320 |
0.89 |
| chr6_119107307_119108229 | 0.41 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
207 |
0.96 |
| chr4_138395338_138395489 | 0.41 |
AB041806 |
hypothetical protein, MNCb-2457 |
215 |
0.85 |
| chr12_18609538_18611003 | 0.40 |
Gm48398 |
predicted gene, 48398 |
114 |
0.96 |
| chr13_54765971_54766190 | 0.40 |
Sncb |
synuclein, beta |
17 |
0.96 |
| chrX_162759829_162760925 | 0.40 |
Rbbp7 |
retinoblastoma binding protein 7, chromatin remodeling factor |
25 |
0.97 |
| chr9_8544024_8544863 | 0.40 |
Trpc6 |
transient receptor potential cation channel, subfamily C, member 6 |
200 |
0.96 |
| chr1_177448201_177448585 | 0.40 |
Zbtb18 |
zinc finger and BTB domain containing 18 |
2572 |
0.22 |
| chr16_14445264_14445415 | 0.40 |
Abcc1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
15669 |
0.17 |
| chr5_28460507_28460658 | 0.40 |
9530036O11Rik |
RIKEN cDNA 9530036O11Rik |
2308 |
0.3 |
| chr13_71963383_71964045 | 0.40 |
Irx1 |
Iroquois homeobox 1 |
9 |
0.98 |
| chr17_83954879_83955189 | 0.39 |
Gm35229 |
predicted gene, 35229 |
1214 |
0.31 |
| chr9_53705535_53706804 | 0.39 |
Rab39 |
RAB39, member RAS oncogene family |
63 |
0.96 |
| chr13_17993545_17993782 | 0.39 |
Yae1d1 |
Yae1 domain containing 1 |
314 |
0.75 |
| chr8_111739568_111739875 | 0.39 |
Bcar1 |
breast cancer anti-estrogen resistance 1 |
4088 |
0.22 |
| chr14_69609343_69609518 | 0.39 |
Loxl2 |
lysyl oxidase-like 2 |
48 |
0.96 |
| chr10_81392422_81392592 | 0.38 |
Smim24 |
small integral membrane protein 24 |
544 |
0.44 |
| chr17_56626470_56627486 | 0.38 |
Lonp1 |
lon peptidase 1, mitochondrial |
91 |
0.92 |
| chr1_64735773_64736678 | 0.38 |
Gm38058 |
predicted gene, 38058 |
15 |
0.96 |
| chr8_90907824_90909226 | 0.38 |
Chd9 |
chromodomain helicase DNA binding protein 9 |
89 |
0.52 |
| chr7_37769955_37770699 | 0.38 |
Zfp536 |
zinc finger protein 536 |
365 |
0.9 |
| chr15_38298982_38300199 | 0.38 |
Klf10 |
Kruppel-like factor 10 |
205 |
0.91 |
| chr7_101821788_101822627 | 0.38 |
Phox2a |
paired-like homeobox 2a |
1461 |
0.21 |
| chr1_120602158_120602362 | 0.37 |
En1 |
engrailed 1 |
158 |
0.96 |
| chr6_89361531_89362433 | 0.37 |
Plxna1 |
plexin A1 |
638 |
0.7 |
| chr12_55598801_55599099 | 0.37 |
Insm2 |
insulinoma-associated 2 |
477 |
0.8 |
| chr7_109703238_109703866 | 0.37 |
Denn2b |
DENN domain containing 2B |
53 |
0.6 |
| chr9_92541870_92542582 | 0.37 |
Plod2 |
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 |
3 |
0.92 |
| chr5_148398815_148400002 | 0.37 |
Slc7a1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
383 |
0.89 |
| chr5_129724468_129725740 | 0.36 |
Nipsnap2 |
nipsnap homolog 2 |
3 |
0.95 |
| chr16_92357164_92357320 | 0.36 |
Kcne1 |
potassium voltage-gated channel, Isk-related subfamily, member 1 |
1632 |
0.26 |
| chr11_117864041_117864389 | 0.36 |
Tmem235 |
transmembrane protein 235 |
3463 |
0.12 |
| chr17_69417243_69417687 | 0.36 |
C030034I22Rik |
RIKEN cDNA C030034I22 gene |
4 |
0.97 |
| chr1_90797381_90797532 | 0.36 |
Col6a3 |
collagen, type VI, alpha 3 |
2735 |
0.29 |
| chr1_25829654_25830461 | 0.36 |
Adgrb3 |
adhesion G protein-coupled receptor B3 |
350 |
0.61 |
| chr8_106893544_106894153 | 0.35 |
Utp4 |
UTP4 small subunit processome component |
197 |
0.4 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.3 | 1.1 | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis(GO:0003253) |
| 0.2 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 0.5 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 0.5 | GO:0033505 | floor plate morphogenesis(GO:0033505) |
| 0.2 | 0.6 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.2 | 0.2 | GO:0072025 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 | 0.9 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.6 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.4 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.1 | 0.2 | GO:0033122 | negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.4 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 | 0.8 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 0.3 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 1.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.4 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.1 | 1.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.1 | 0.2 | GO:0071655 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.1 | 0.5 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.5 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.3 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.4 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.5 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.2 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.2 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.1 | 0.3 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.2 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.2 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:1903011 | negative regulation of bone development(GO:1903011) |
| 0.1 | 0.4 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) |
| 0.1 | 0.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.7 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.3 | GO:0019230 | proprioception(GO:0019230) |
| 0.1 | 0.3 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.2 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.7 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.3 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.2 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.7 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.1 | 0.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.5 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.2 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.8 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.8 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.2 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.0 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.4 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.7 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.0 | 0.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 1.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.0 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.0 | 0.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.0 | 0.4 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.2 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.4 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0035788 | cell migration involved in kidney development(GO:0035787) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.1 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.2 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.4 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.4 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.0 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 0.0 | 0.4 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 | 0.0 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.2 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.6 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.1 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.4 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.0 | 0.2 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.0 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.0 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.0 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.2 | GO:0071475 | cellular response to salt stress(GO:0071472) cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.0 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.0 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.1 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.0 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.0 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.0 | 0.0 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0086011 | membrane repolarization during action potential(GO:0086011) |
| 0.0 | 0.0 | GO:0042505 | tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.0 | 0.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.0 | GO:0044845 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.2 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.0 | 0.0 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0045180 | basal cortex(GO:0045180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 1.0 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 0.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 0.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.5 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.1 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 0.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.3 | GO:0015189 | arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.5 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 1.4 | GO:0043738 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.3 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.3 | GO:0043912 | D-lysine oxidase activity(GO:0043912) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0043338 | CTP:2,3-di-O-geranylgeranyl-sn-glycero-1-phosphate cytidyltransferase activity(GO:0043338) phospholactate guanylyltransferase activity(GO:0043814) ATP:coenzyme F420 adenylyltransferase activity(GO:0043910) UDP-N-acetylgalactosamine diphosphorylase activity(GO:0052630) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.4 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.1 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |