Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
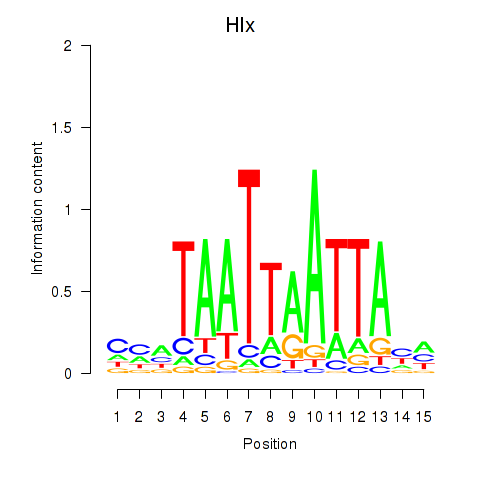
Results for Hlx
Z-value: 0.82
Transcription factors associated with Hlx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hlx
|
ENSMUSG00000039377.6 | H2.0-like homeobox |
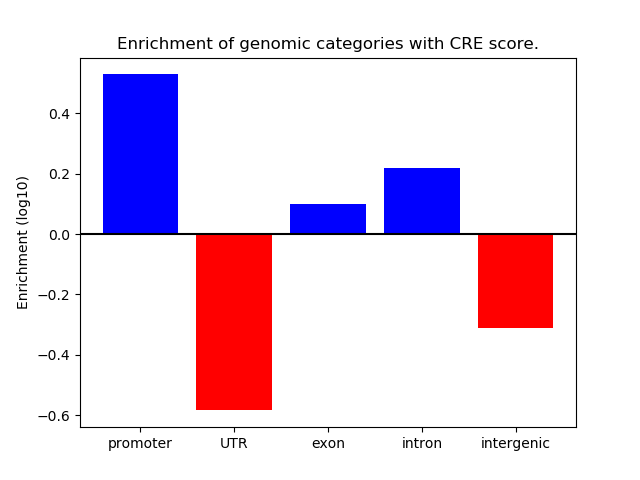
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
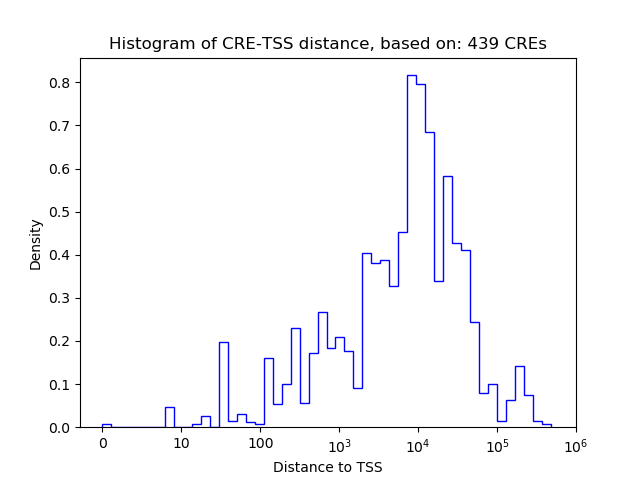
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_184727788_184727992 | Hlx | 3708 | 0.171022 | -0.24 | 7.7e-02 | Click! |
| chr1_184731813_184732486 | Hlx | 470 | 0.749555 | -0.15 | 2.9e-01 | Click! |
| chr1_184727523_184727685 | Hlx | 3994 | 0.166076 | -0.14 | 3.0e-01 | Click! |
| chr1_184732596_184733212 | Hlx | 285 | 0.870881 | 0.09 | 5.0e-01 | Click! |
| chr1_184733319_184733470 | Hlx | 775 | 0.552845 | -0.03 | 8.5e-01 | Click! |
Activity of the Hlx motif across conditions
Conditions sorted by the z-value of the Hlx motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
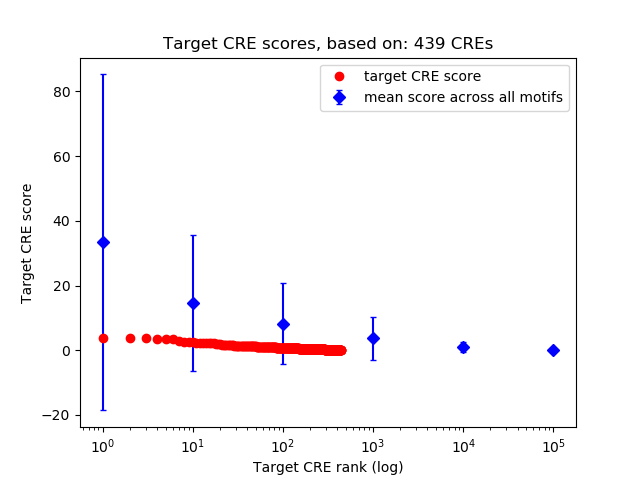
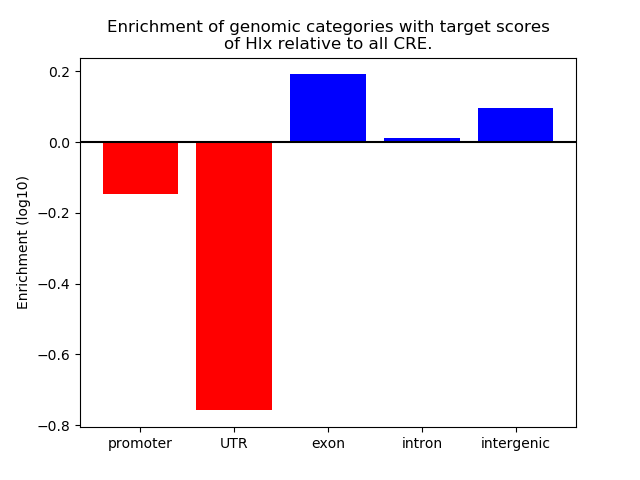
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_117897075_117897226 | 3.87 |
4933411E08Rik |
RIKEN cDNA 4933411E08 gene |
28309 |
0.12 |
| chr2_119566179_119566455 | 3.76 |
Chp1 |
calcineurin-like EF hand protein 1 |
288 |
0.85 |
| chr1_177475904_177476057 | 3.63 |
Gm37306 |
predicted gene, 37306 |
8602 |
0.17 |
| chrX_93831605_93832731 | 3.58 |
Pdk3 |
pyruvate dehydrogenase kinase, isoenzyme 3 |
33 |
0.98 |
| chr19_43767999_43768210 | 3.53 |
Cutc |
cutC copper transporter |
3231 |
0.18 |
| chr4_117136303_117136464 | 3.49 |
Plk3 |
polo like kinase 3 |
2420 |
0.1 |
| chr16_78040391_78040542 | 2.74 |
Gm25025 |
predicted gene, 25025 |
10251 |
0.23 |
| chr10_69253249_69253618 | 2.40 |
Rhobtb1 |
Rho-related BTB domain containing 1 |
12204 |
0.19 |
| chr13_62952253_62952404 | 2.40 |
Gm48812 |
predicted gene, 48812 |
2886 |
0.22 |
| chr4_49451596_49451772 | 2.39 |
Acnat1 |
acyl-coenzyme A amino acid N-acyltransferase 1 |
547 |
0.69 |
| chr13_47111196_47111529 | 2.37 |
1700026N04Rik |
RIKEN cDNA 1700026N04 gene |
4943 |
0.13 |
| chr13_93234781_93234932 | 2.25 |
Tent2 |
terminal nucleotidyltransferase 2 |
42471 |
0.12 |
| chr4_14273616_14273932 | 2.15 |
Gm24908 |
predicted gene, 24908 |
34156 |
0.18 |
| chr3_108771125_108771338 | 2.14 |
Aknad1 |
AKNA domain containing 1 |
11017 |
0.15 |
| chr11_4095576_4095728 | 2.11 |
Mtfp1 |
mitochondrial fission process 1 |
207 |
0.87 |
| chr3_34504199_34504577 | 2.10 |
Gm29135 |
predicted gene 29135 |
22181 |
0.16 |
| chr15_98608664_98610204 | 2.09 |
Adcy6 |
adenylate cyclase 6 |
598 |
0.55 |
| chr7_45103031_45103408 | 2.04 |
Fcgrt |
Fc receptor, IgG, alpha chain transporter |
133 |
0.83 |
| chr10_125785483_125786054 | 2.02 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
180400 |
0.03 |
| chr5_136739184_136739335 | 1.86 |
Col26a1 |
collagen, type XXVI, alpha 1 |
5290 |
0.23 |
| chrX_10877865_10878215 | 1.71 |
Gm14473 |
predicted gene 14473 |
12407 |
0.26 |
| chr13_36202916_36203067 | 1.63 |
Gm48765 |
predicted gene, 48765 |
13268 |
0.16 |
| chr1_59212186_59212347 | 1.59 |
Als2 |
alsin Rho guanine nucleotide exchange factor |
6860 |
0.17 |
| chr10_120387024_120387466 | 1.57 |
9230105E05Rik |
RIKEN cDNA 9230105E05 gene |
2278 |
0.28 |
| chr16_44672098_44672717 | 1.57 |
Nepro |
nucleolus and neural progenitor protein |
51894 |
0.11 |
| chr8_125782363_125782518 | 1.54 |
Pcnx2 |
pecanex homolog 2 |
8312 |
0.24 |
| chr13_101427845_101427996 | 1.52 |
Gm36994 |
predicted gene, 36994 |
15331 |
0.2 |
| chr1_162020499_162020732 | 1.49 |
2810442N19Rik |
RIKEN cDNA 2810442N19 gene |
15443 |
0.14 |
| chr7_75761095_75761254 | 1.43 |
Gm23251 |
predicted gene, 23251 |
3806 |
0.19 |
| chr9_113833592_113833892 | 1.42 |
Clasp2 |
CLIP associating protein 2 |
21142 |
0.2 |
| chr4_132072343_132072494 | 1.39 |
Epb41 |
erythrocyte membrane protein band 4.1 |
321 |
0.8 |
| chr11_78401648_78401948 | 1.39 |
Slc13a2os |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2, opposite strand |
7313 |
0.1 |
| chr2_170091681_170092036 | 1.37 |
Zfp217 |
zinc finger protein 217 |
39362 |
0.2 |
| chr13_63039019_63039172 | 1.36 |
Aopep |
aminopeptidase O |
23602 |
0.15 |
| chr4_154359614_154359823 | 1.33 |
Prdm16 |
PR domain containing 16 |
11261 |
0.18 |
| chr1_36670453_36670635 | 1.32 |
Gm24133 |
predicted gene, 24133 |
6579 |
0.12 |
| chr14_7896790_7896982 | 1.30 |
Flnb |
filamin, beta |
12145 |
0.19 |
| chr17_56036249_56036917 | 1.26 |
Sh3gl1 |
SH3-domain GRB2-like 1 |
7 |
0.94 |
| chr14_21462396_21462784 | 1.25 |
Gm30363 |
predicted gene, 30363 |
8831 |
0.19 |
| chr2_60142673_60142824 | 1.25 |
Ldha-ps |
lactate dehydrogenase A, pseudogene |
8693 |
0.16 |
| chr19_24534375_24534662 | 1.22 |
Pip5k1b |
phosphatidylinositol-4-phosphate 5-kinase, type 1 beta |
21271 |
0.17 |
| chr14_122909784_122909949 | 1.22 |
4930594M22Rik |
RIKEN cDNA 4930594M22 gene |
3164 |
0.2 |
| chr9_32545148_32545570 | 1.21 |
Fli1 |
Friend leukemia integration 1 |
2498 |
0.19 |
| chr13_14040175_14040467 | 1.21 |
Tbce |
tubulin-specific chaperone E |
683 |
0.47 |
| chr6_71130459_71130610 | 1.20 |
Thnsl2 |
threonine synthase-like 2 (bacterial) |
1438 |
0.31 |
| chr8_110022869_110023026 | 1.20 |
Gm45888 |
predicted gene 45888 |
8623 |
0.12 |
| chr7_75863314_75863477 | 1.20 |
Klhl25 |
kelch-like 25 |
14954 |
0.22 |
| chr10_53101557_53101708 | 1.19 |
Gm47622 |
predicted gene, 47622 |
911 |
0.65 |
| chr2_85172390_85172701 | 1.17 |
Gm13713 |
predicted gene 13713 |
11767 |
0.11 |
| chr13_43480925_43481874 | 1.17 |
Ranbp9 |
RAN binding protein 9 |
117 |
0.95 |
| chr11_3170338_3170795 | 1.13 |
Sfi1 |
Sfi1 homolog, spindle assembly associated (yeast) |
138 |
0.93 |
| chr4_97601207_97601387 | 1.13 |
E130114P18Rik |
RIKEN cDNA E130114P18 gene |
16661 |
0.23 |
| chr14_19707913_19708072 | 1.13 |
Gm49341 |
predicted gene, 49341 |
10783 |
0.13 |
| chr5_21264473_21264673 | 1.10 |
Gsap |
gamma-secretase activating protein |
13414 |
0.16 |
| chr18_56871340_56871513 | 1.10 |
Gm18087 |
predicted gene, 18087 |
44662 |
0.14 |
| chr4_137481273_137481486 | 1.09 |
Hspg2 |
perlecan (heparan sulfate proteoglycan 2) |
12576 |
0.13 |
| chr10_71338295_71338446 | 1.08 |
Cisd1 |
CDGSH iron sulfur domain 1 |
6584 |
0.14 |
| chr13_62888714_62888865 | 1.06 |
Fbp1 |
fructose bisphosphatase 1 |
507 |
0.73 |
| chr5_103753230_103754272 | 1.06 |
Aff1 |
AF4/FMR2 family, member 1 |
411 |
0.87 |
| chr12_47466088_47466239 | 1.05 |
Gm48578 |
predicted gene, 48578 |
66094 |
0.15 |
| chr15_102753205_102753442 | 1.02 |
Calcoco1 |
calcium binding and coiled coil domain 1 |
31145 |
0.11 |
| chr2_167713774_167713925 | 1.02 |
A530013C23Rik |
RIKEN cDNA A530013C23 gene |
22668 |
0.1 |
| chr16_45924188_45924367 | 1.01 |
Gm15640 |
predicted gene 15640 |
12046 |
0.17 |
| chr15_7625219_7625370 | 1.01 |
Gm37743 |
predicted gene, 37743 |
45046 |
0.15 |
| chr11_69418367_69419708 | 0.99 |
Kdm6b |
KDM1 lysine (K)-specific demethylase 6B |
5362 |
0.09 |
| chr1_84993310_84993634 | 0.99 |
Gm29284 |
predicted gene 29284 |
440 |
0.72 |
| chr2_151976038_151976372 | 0.98 |
Fam110a |
family with sequence similarity 110, member A |
2239 |
0.21 |
| chr6_143142224_143142402 | 0.98 |
Gm44306 |
predicted gene, 44306 |
7226 |
0.14 |
| chr2_155894841_155894992 | 0.98 |
Uqcc1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
15688 |
0.12 |
| chr8_94898997_94899206 | 0.97 |
Ccdc102a |
coiled-coil domain containing 102A |
18420 |
0.1 |
| chr4_137858517_137858672 | 0.96 |
Ece1 |
endothelin converting enzyme 1 |
3643 |
0.24 |
| chr16_25293836_25294037 | 0.96 |
Tprg |
transformation related protein 63 regulated |
7115 |
0.32 |
| chr6_140147796_140147947 | 0.95 |
Gm44159 |
predicted gene, 44159 |
15804 |
0.17 |
| chr15_75863041_75863192 | 0.93 |
Gsdmd |
gasdermin D |
737 |
0.46 |
| chr2_109995179_109995348 | 0.92 |
Lgr4 |
leucine-rich repeat-containing G protein-coupled receptor 4 |
5265 |
0.22 |
| chr4_142049656_142049814 | 0.91 |
Gm13053 |
predicted gene 13053 |
1157 |
0.39 |
| chr1_85615441_85615607 | 0.90 |
Gm10552 |
predicted gene 10552 |
3628 |
0.12 |
| chr6_25664249_25664412 | 0.90 |
Gpr37 |
G protein-coupled receptor 37 |
25462 |
0.25 |
| chr1_66922212_66922363 | 0.90 |
Myl1 |
myosin, light polypeptide 1 |
12411 |
0.12 |
| chr2_171734745_171734896 | 0.86 |
1700028P15Rik |
RIKEN cDNA 1700028P15 gene |
227311 |
0.02 |
| chr1_88491666_88491830 | 0.86 |
Glrp1 |
glutamine repeat protein 1 |
18318 |
0.14 |
| chr9_63201505_63201702 | 0.84 |
Skor1 |
SKI family transcriptional corepressor 1 |
52642 |
0.12 |
| chr2_18586936_18587492 | 0.84 |
Gm13353 |
predicted gene 13353 |
19088 |
0.18 |
| chr14_16962435_16962707 | 0.82 |
Gm18078 |
predicted gene, 18078 |
8965 |
0.24 |
| chr11_62605415_62606716 | 0.82 |
Lrrc75aos2 |
leucine rich repeat containing 75A, opposite strand 2 |
298 |
0.72 |
| chr1_80408527_80409167 | 0.82 |
Gm6189 |
predicted gene 6189 |
23662 |
0.13 |
| chr7_100559420_100559571 | 0.81 |
Mrpl48 |
mitochondrial ribosomal protein L48 |
11875 |
0.09 |
| chr1_45767537_45767744 | 0.81 |
Gm8307 |
predicted gene 8307 |
697 |
0.6 |
| chr7_6781231_6781382 | 0.81 |
Mir3099 |
microRNA 3099 |
22281 |
0.09 |
| chr7_137306238_137306389 | 0.80 |
Ebf3 |
early B cell factor 3 |
7603 |
0.2 |
| chr17_25127767_25127933 | 0.79 |
Clcn7 |
chloride channel, voltage-sensitive 7 |
5541 |
0.1 |
| chr9_50501327_50501514 | 0.79 |
Plet1os |
placenta expressed transcript 1, opposite strand |
3385 |
0.16 |
| chr1_176995106_176995337 | 0.78 |
Sdccag8 |
serologically defined colon cancer antigen 8 |
2788 |
0.21 |
| chr1_85032083_85032289 | 0.77 |
AC167036.2 |
novel protein |
3342 |
0.13 |
| chr5_151046930_151047128 | 0.77 |
Gm8675 |
predicted gene 8675 |
37728 |
0.16 |
| chrX_38473528_38474265 | 0.77 |
Gm7598 |
predicted gene 7598 |
2565 |
0.27 |
| chr14_17786377_17786528 | 0.77 |
Gm48320 |
predicted gene, 48320 |
15330 |
0.28 |
| chr2_172261099_172261318 | 0.75 |
Mc3r |
melanocortin 3 receptor |
12716 |
0.17 |
| chr3_28060860_28061033 | 0.75 |
Pld1 |
phospholipase D1 |
8568 |
0.28 |
| chr2_170158021_170158172 | 0.74 |
Zfp217 |
zinc finger protein 217 |
9993 |
0.28 |
| chr1_98147453_98147604 | 0.74 |
1810006J02Rik |
RIKEN cDNA 1810006J02 gene |
16061 |
0.18 |
| chr7_101837492_101838185 | 0.73 |
Inppl1 |
inositol polyphosphate phosphatase-like 1 |
35 |
0.49 |
| chr1_130732805_130732969 | 0.73 |
AA986860 |
expressed sequence AA986860 |
777 |
0.43 |
| chr10_128336232_128336487 | 0.72 |
Cs |
citrate synthase |
1375 |
0.18 |
| chr10_20046153_20046446 | 0.72 |
Map3k5 |
mitogen-activated protein kinase kinase kinase 5 |
53226 |
0.13 |
| chr3_115660838_115661123 | 0.72 |
S1pr1 |
sphingosine-1-phosphate receptor 1 |
54092 |
0.13 |
| chr4_106909039_106909761 | 0.71 |
Ssbp3 |
single-stranded DNA binding protein 3 |
1301 |
0.48 |
| chr14_117674551_117674852 | 0.70 |
Mir6239 |
microRNA 6239 |
279146 |
0.01 |
| chr1_166002288_166003185 | 0.69 |
Pou2f1 |
POU domain, class 2, transcription factor 1 |
58 |
0.72 |
| chr12_75826912_75827184 | 0.69 |
Syne2 |
spectrin repeat containing, nuclear envelope 2 |
8698 |
0.25 |
| chr11_4553637_4553951 | 0.68 |
Mtmr3 |
myotubularin related protein 3 |
7526 |
0.17 |
| chr1_192823078_192823550 | 0.66 |
Gm38360 |
predicted gene, 38360 |
2187 |
0.22 |
| chr12_103969871_103970197 | 0.65 |
Serpina1e |
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
11059 |
0.1 |
| chr6_134147989_134148473 | 0.64 |
Gm17088 |
predicted gene 17088 |
23776 |
0.18 |
| chr7_65803210_65803361 | 0.64 |
1810008I18Rik |
RIKEN cDNA 1810008I18 gene |
174 |
0.95 |
| chr1_64087943_64089121 | 0.63 |
Gm13748 |
predicted gene 13748 |
9878 |
0.19 |
| chr17_47914586_47915244 | 0.63 |
Gm15556 |
predicted gene 15556 |
7463 |
0.14 |
| chr10_18370345_18371012 | 0.63 |
Nhsl1 |
NHS-like 1 |
19644 |
0.19 |
| chr3_60500259_60500437 | 0.62 |
Mbnl1 |
muscleblind like splicing factor 1 |
587 |
0.8 |
| chr2_93008105_93008300 | 0.62 |
Prdm11 |
PR domain containing 11 |
5708 |
0.22 |
| chr11_105549454_105549608 | 0.62 |
Tanc2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
40455 |
0.15 |
| chr7_19119292_19119545 | 0.61 |
Fbxo46 |
F-box protein 46 |
441 |
0.56 |
| chr13_22020931_22021318 | 0.61 |
Gm11290 |
predicted gene 11290 |
3618 |
0.06 |
| chr12_109552974_109553125 | 0.61 |
Meg3 |
maternally expressed 3 |
3794 |
0.05 |
| chr6_89291609_89291772 | 0.61 |
4930512J16Rik |
RIKEN cDNA 4930512J16 gene |
5965 |
0.14 |
| chr2_34825635_34825786 | 0.60 |
Fbxw2 |
F-box and WD-40 domain protein 2 |
312 |
0.82 |
| chr17_78327187_78327654 | 0.60 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
23469 |
0.16 |
| chr19_9924707_9924858 | 0.59 |
Gm36913 |
predicted gene, 36913 |
3752 |
0.1 |
| chr17_78273911_78274066 | 0.59 |
Crim1 |
cysteine rich transmembrane BMP regulator 1 (chordin like) |
6195 |
0.2 |
| chr12_52671451_52671886 | 0.58 |
Akap6 |
A kinase (PRKA) anchor protein 6 |
27715 |
0.15 |
| chr12_37142164_37142324 | 0.58 |
Meox2 |
mesenchyme homeobox 2 |
33704 |
0.16 |
| chr8_104365772_104365923 | 0.58 |
Gm45877 |
predicted gene 45877 |
2179 |
0.16 |
| chr2_178610675_178610841 | 0.57 |
Cdh26 |
cadherin-like 26 |
150128 |
0.04 |
| chr11_53818750_53818901 | 0.57 |
Gm12216 |
predicted gene 12216 |
22950 |
0.11 |
| chr18_23748563_23748754 | 0.57 |
Gm15972 |
predicted gene 15972 |
3647 |
0.2 |
| chr3_95517252_95517598 | 0.56 |
Ctss |
cathepsin S |
9361 |
0.1 |
| chr11_98925326_98925710 | 0.56 |
Rara |
retinoic acid receptor, alpha |
2300 |
0.17 |
| chr12_54985296_54986516 | 0.56 |
Baz1a |
bromodomain adjacent to zinc finger domain 1A |
30 |
0.91 |
| chr18_5002577_5002904 | 0.55 |
Svil |
supervillin |
7804 |
0.3 |
| chr3_129296851_129297002 | 0.55 |
Gm42628 |
predicted gene 42628 |
11657 |
0.16 |
| chr14_65737090_65737241 | 0.55 |
Scara5 |
scavenger receptor class A, member 5 |
60688 |
0.1 |
| chr11_29547325_29547986 | 0.54 |
Rps27a |
ribosomal protein S27A |
205 |
0.59 |
| chr4_132874359_132874577 | 0.54 |
Stx12 |
syntaxin 12 |
10041 |
0.12 |
| chr17_30611617_30611888 | 0.54 |
Glo1 |
glyoxalase 1 |
817 |
0.45 |
| chr18_78350420_78350571 | 0.54 |
Gm6133 |
predicted gene 6133 |
741 |
0.78 |
| chr12_112333322_112333806 | 0.53 |
Gm38123 |
predicted gene, 38123 |
16503 |
0.18 |
| chr13_95777279_95777613 | 0.53 |
Iqgap2 |
IQ motif containing GTPase activating protein 2 |
13049 |
0.19 |
| chr12_105757874_105758401 | 0.53 |
Ak7 |
adenylate kinase 7 |
12862 |
0.17 |
| chrX_96134190_96134604 | 0.53 |
Gm24718 |
predicted gene, 24718 |
16575 |
0.18 |
| chr1_180179935_180180086 | 0.53 |
Coq8a |
coenzyme Q8A |
1088 |
0.44 |
| chr10_115263315_115263511 | 0.53 |
Gm8942 |
predicted gene 8942 |
6565 |
0.17 |
| chr1_136911165_136911329 | 0.53 |
Nr5a2 |
nuclear receptor subfamily 5, group A, member 2 |
29336 |
0.18 |
| chr4_43598778_43598951 | 0.52 |
Gm12472 |
predicted gene 12472 |
10792 |
0.07 |
| chr13_37859595_37859751 | 0.52 |
Rreb1 |
ras responsive element binding protein 1 |
1739 |
0.39 |
| chr2_174291748_174291899 | 0.52 |
Gnasas1 |
GNAS antisense RNA 1 |
3566 |
0.16 |
| chr11_47188652_47188809 | 0.52 |
Sgcd |
sarcoglycan, delta (dystrophin-associated glycoprotein) |
190792 |
0.03 |
| chr17_86301671_86301829 | 0.51 |
2010106C02Rik |
RIKEN cDNA 2010106C02 gene |
14572 |
0.27 |
| chr12_79532095_79532284 | 0.51 |
Rad51b |
RAD51 paralog B |
204836 |
0.02 |
| chr2_167902375_167902526 | 0.51 |
Ptpn1 |
protein tyrosine phosphatase, non-receptor type 1 |
29607 |
0.14 |
| chr10_127355403_127355866 | 0.50 |
Inhbe |
inhibin beta-E |
1223 |
0.25 |
| chr18_74854672_74855018 | 0.50 |
Gm24559 |
predicted gene, 24559 |
51589 |
0.08 |
| chr16_95924762_95924926 | 0.50 |
1600002D24Rik |
RIKEN cDNA 1600002D24 gene |
4233 |
0.21 |
| chr13_42342256_42342434 | 0.50 |
Gm47118 |
predicted gene, 47118 |
37354 |
0.17 |
| chr12_117250438_117250838 | 0.49 |
Mir153 |
microRNA 153 |
179 |
0.97 |
| chr2_131254315_131254509 | 0.49 |
Pank2 |
pantothenate kinase 2 |
8083 |
0.11 |
| chr6_144902234_144902547 | 0.49 |
Gm22792 |
predicted gene, 22792 |
98758 |
0.06 |
| chr5_25064693_25064847 | 0.49 |
Prkag2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
35872 |
0.13 |
| chr1_151580285_151580566 | 0.48 |
Fam129a |
family with sequence similarity 129, member A |
8929 |
0.17 |
| chr17_56688104_56688477 | 0.48 |
Ranbp3 |
RAN binding protein 3 |
8445 |
0.11 |
| chr2_134350455_134350845 | 0.48 |
Hao1 |
hydroxyacid oxidase 1, liver |
203657 |
0.03 |
| chr15_64026966_64027117 | 0.48 |
Gm25628 |
predicted gene, 25628 |
1093 |
0.51 |
| chr5_15487606_15487855 | 0.48 |
Gm21149 |
predicted gene, 21149 |
11207 |
0.16 |
| chr7_35116059_35116210 | 0.48 |
Cebpa |
CCAAT/enhancer binding protein (C/EBP), alpha |
3159 |
0.12 |
| chr3_55075221_55075624 | 0.48 |
Gm43555 |
predicted gene 43555 |
17766 |
0.13 |
| chr11_22348041_22348563 | 0.47 |
Ehbp1 |
EH domain binding protein 1 |
6010 |
0.25 |
| chr9_104071426_104071577 | 0.47 |
Acad11 |
acyl-Coenzyme A dehydrogenase family, member 11 |
7768 |
0.1 |
| chr9_69291313_69291464 | 0.47 |
Rora |
RAR-related orphan receptor alpha |
1706 |
0.45 |
| chr1_17655681_17656007 | 0.47 |
Gm28154 |
predicted gene 28154 |
14763 |
0.2 |
| chr17_84777000_84777216 | 0.46 |
Lrpprc |
leucine-rich PPR-motif containing |
435 |
0.81 |
| chr11_105125823_105126961 | 0.46 |
Mettl2 |
methyltransferase like 2 |
33 |
0.97 |
| chr8_109567958_109568109 | 0.46 |
Txnl4b |
thioredoxin-like 4B |
855 |
0.48 |
| chr9_79869537_79869690 | 0.46 |
Gm20477 |
predicted gene 20477 |
31226 |
0.13 |
| chr16_36707794_36708006 | 0.46 |
Ildr1 |
immunoglobulin-like domain containing receptor 1 |
7638 |
0.14 |
| chr8_64746788_64747015 | 0.45 |
Klhl2 |
kelch-like 2, Mayven |
8070 |
0.16 |
| chr8_25772876_25773027 | 0.45 |
Bag4 |
BCL2-associated athanogene 4 |
4631 |
0.11 |
| chr10_102895997_102896156 | 0.45 |
Gm5175 |
predicted gene 5175 |
76705 |
0.1 |
| chr4_141734704_141734869 | 0.45 |
Ddi2 |
DNA-damage inducible protein 2 |
11367 |
0.12 |
| chr19_14596815_14596966 | 0.44 |
Tle4 |
transducin-like enhancer of split 4 |
1161 |
0.62 |
| chr1_54913082_54913233 | 0.44 |
A130048G24Rik |
RIKEN cDNA A130048G24 gene |
3437 |
0.24 |
| chr14_41115323_41115474 | 0.44 |
Mat1a |
methionine adenosyltransferase I, alpha |
10017 |
0.12 |
| chr15_58796543_58796694 | 0.44 |
Gm20712 |
predicted gene 20712 |
14667 |
0.14 |
| chr4_74401736_74401893 | 0.43 |
Kdm4c |
lysine (K)-specific demethylase 4C |
139004 |
0.04 |
| chr16_52446109_52446270 | 0.43 |
Alcam |
activated leukocyte cell adhesion molecule |
6276 |
0.33 |
| chr13_80623585_80623746 | 0.43 |
Gm46388 |
predicted gene, 46388 |
130656 |
0.05 |
| chr9_122074963_122075114 | 0.43 |
Gm39465 |
predicted gene, 39465 |
23575 |
0.1 |
| chr19_55110604_55110837 | 0.42 |
Gpam |
glycerol-3-phosphate acyltransferase, mitochondrial |
7629 |
0.21 |
| chr4_146482529_146482708 | 0.41 |
Gm13245 |
predicted gene 13245 |
1951 |
0.25 |
| chr1_136228974_136229345 | 0.41 |
Inava |
innate immunity activator |
860 |
0.42 |
| chr2_167799405_167799696 | 0.40 |
9230111E07Rik |
RIKEN cDNA 9230111E07 gene |
17497 |
0.14 |
| chr9_115243693_115243977 | 0.40 |
Stmn1-rs1 |
stathmin 1, related sequence 1 |
38146 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.8 | 3.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.2 | 1.0 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.2 | 0.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 1.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.1 | 0.4 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.2 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.1 | 0.4 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.4 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.5 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 2.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.4 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.2 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.3 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.6 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.0 | GO:2000564 | regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 1.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.5 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 1.9 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.2 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.2 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.0 | 0.2 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.3 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 1.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.1 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.3 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.0 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0042891 | antibiotic transport(GO:0042891) |
| 0.0 | 0.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.0 | GO:0071926 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.0 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 1.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.3 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.3 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.0 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.0 | GO:0048208 | trophectodermal cellular morphogenesis(GO:0001831) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.0 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0071651 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.0 | GO:2001260 | regulation of semaphorin-plexin signaling pathway(GO:2001260) |
| 0.0 | 0.0 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 | 0.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.0 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.0 | GO:0015677 | copper ion import(GO:0015677) ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.0 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.3 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 1.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.4 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 1.3 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 3.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 1.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.6 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.6 | 1.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 1.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.4 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 1.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.8 | GO:0018031 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.1 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 3.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.3 | GO:0008758 | thiamine-pyrophosphatase activity(GO:0004787) UDP-2,3-diacylglucosamine hydrolase activity(GO:0008758) dATP pyrophosphohydrolase activity(GO:0008828) dihydroneopterin monophosphate phosphatase activity(GO:0019176) dihydroneopterin triphosphate pyrophosphohydrolase activity(GO:0019177) dTTP diphosphatase activity(GO:0036218) phosphocholine hydrolase activity(GO:0044606) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 2.4 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.0 | GO:1990188 | euchromatin binding(GO:1990188) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 1.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |