Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
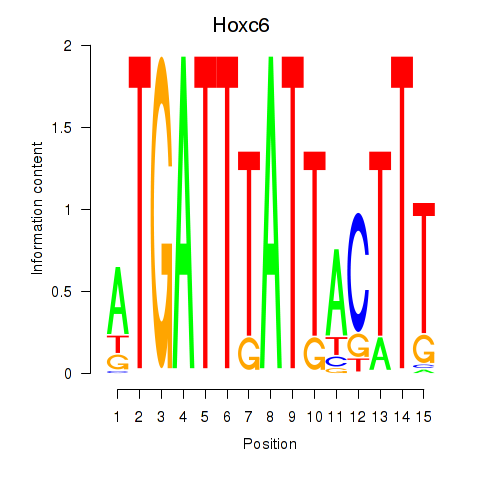
Results for Hoxc6
Z-value: 1.43
Transcription factors associated with Hoxc6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxc6
|
ENSMUSG00000001661.4 | homeobox C6 |
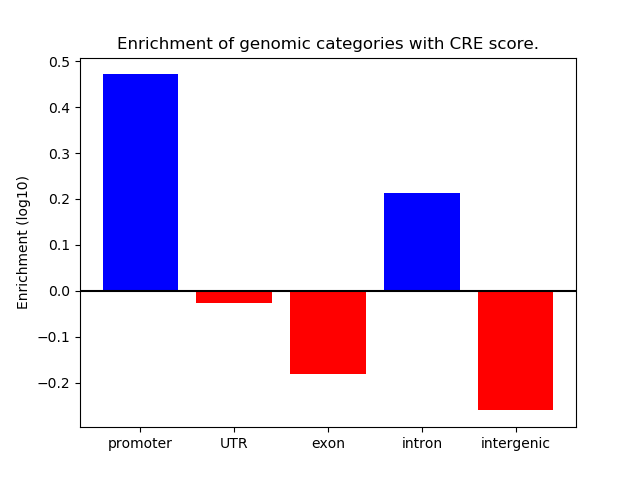
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_103009878_103010029 | Hoxc6 | 380 | 0.695759 | 0.78 | 1.9e-12 | Click! |
| chr15_102997941_102998298 | Hoxc6 | 138 | 0.899628 | 0.77 | 8.1e-12 | Click! |
| chr15_102998391_102998542 | Hoxc6 | 209 | 0.849981 | 0.72 | 7.1e-10 | Click! |
| chr15_102999617_102999826 | Hoxc6 | 808 | 0.387453 | 0.72 | 8.3e-10 | Click! |
| chr15_103000513_103000885 | Hoxc6 | 170 | 0.879888 | 0.71 | 1.8e-09 | Click! |
Activity of the Hoxc6 motif across conditions
Conditions sorted by the z-value of the Hoxc6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
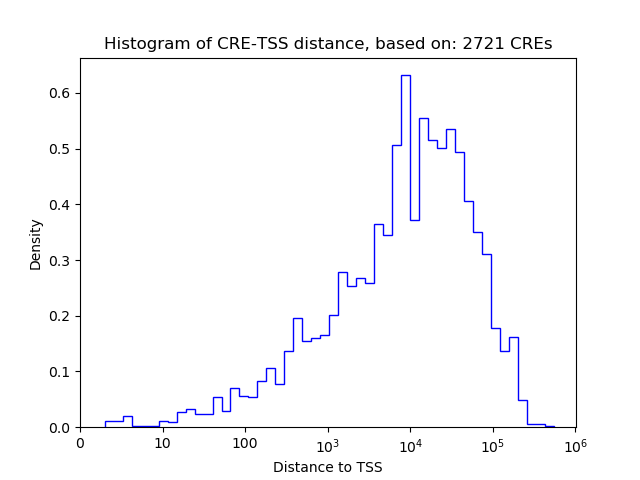
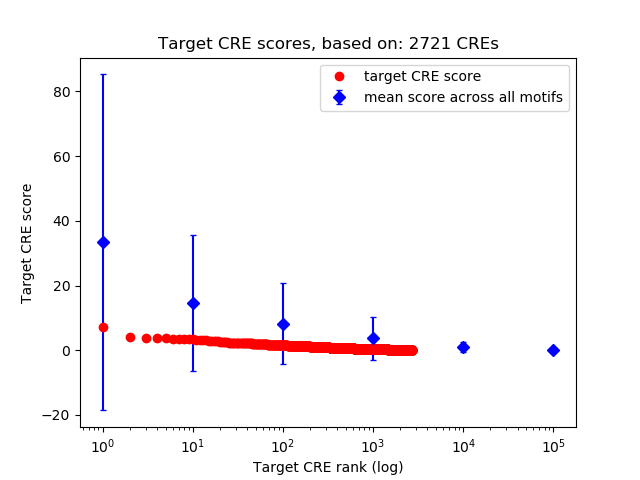
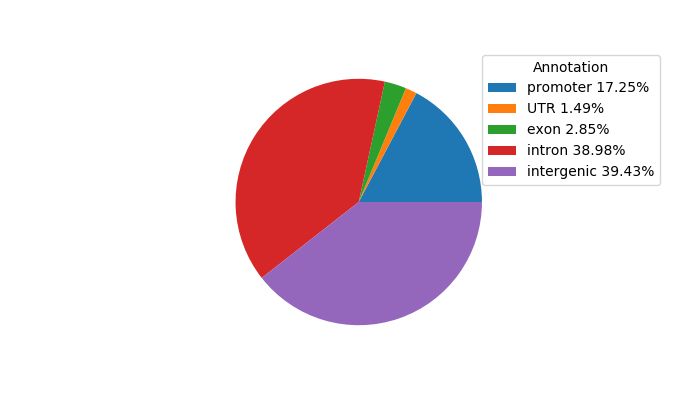
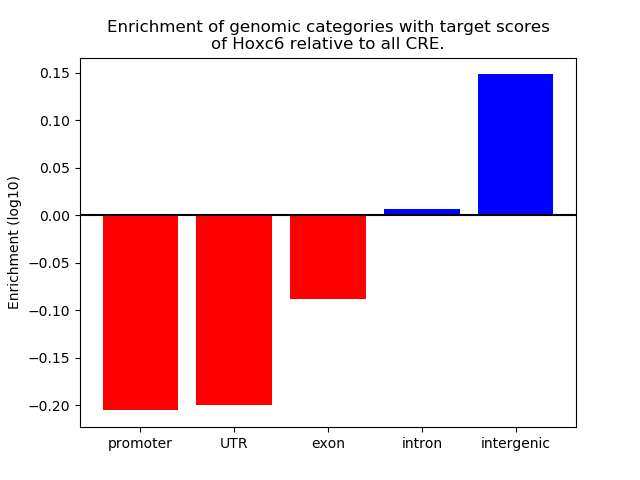
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_70697879_70698218 | 7.26 |
Gm50261 |
predicted gene, 50261 |
8449 |
0.22 |
| chr11_45389248_45390018 | 4.03 |
Gm12162 |
predicted gene 12162 |
31151 |
0.21 |
| chr18_30378314_30378510 | 3.87 |
Gm7926 |
predicted gene 7926 |
41648 |
0.14 |
| chr8_53171807_53171969 | 3.68 |
Gm23666 |
predicted gene, 23666 |
165480 |
0.04 |
| chr9_23037506_23037746 | 3.63 |
Bmper |
BMP-binding endothelial regulator |
185450 |
0.03 |
| chr9_89496365_89496556 | 3.56 |
Gm47403 |
predicted gene, 47403 |
63924 |
0.11 |
| chr5_131585577_131586049 | 3.49 |
Gm27266 |
predicted gene, 27266 |
7826 |
0.13 |
| chr2_171734745_171734896 | 3.46 |
1700028P15Rik |
RIKEN cDNA 1700028P15 gene |
227311 |
0.02 |
| chr15_103061553_103061747 | 3.37 |
5730585A16Rik |
RIKEN cDNA 5730585A16 gene |
2344 |
0.16 |
| chr5_45282398_45282967 | 3.36 |
Gm43303 |
predicted gene 43303 |
30753 |
0.17 |
| chr7_3410706_3410863 | 3.22 |
Gm23450 |
predicted gene, 23450 |
4282 |
0.1 |
| chr9_15964185_15964748 | 3.16 |
Fat3 |
FAT atypical cadherin 3 |
6644 |
0.29 |
| chr8_61273669_61273862 | 3.09 |
1700001D01Rik |
RIKEN cDNA 1700001D01 gene |
15107 |
0.2 |
| chr2_74721103_74721332 | 3.03 |
Hoxd4 |
homeobox D4 |
761 |
0.32 |
| chr14_22783336_22783559 | 2.93 |
Gm7473 |
predicted gene 7473 |
8203 |
0.31 |
| chr11_96333479_96333781 | 2.90 |
Hoxb3 |
homeobox B3 |
5568 |
0.08 |
| chr2_20990174_20990336 | 2.89 |
Gm13375 |
predicted gene 13375 |
21201 |
0.18 |
| chr14_23638190_23638341 | 2.87 |
Kcnma1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
11924 |
0.27 |
| chr2_109991911_109992079 | 2.72 |
Lgr4 |
leucine-rich repeat-containing G protein-coupled receptor 4 |
8533 |
0.2 |
| chr14_67283363_67283593 | 2.61 |
Gm23178 |
predicted gene, 23178 |
8515 |
0.17 |
| chr2_48318409_48318876 | 2.60 |
Gm13482 |
predicted gene 13482 |
19734 |
0.23 |
| chr1_125013472_125013798 | 2.52 |
Dpp10 |
dipeptidylpeptidase 10 |
167596 |
0.04 |
| chr13_4192626_4192777 | 2.42 |
Akr1c13 |
aldo-keto reductase family 1, member C13 |
1157 |
0.38 |
| chr2_110379919_110380070 | 2.41 |
Fibin |
fin bud initiation factor homolog (zebrafish) |
16811 |
0.21 |
| chr14_22216025_22216202 | 2.35 |
Lrmda |
leucine rich melanocyte differentiation associated |
94052 |
0.08 |
| chr8_26594087_26594265 | 2.35 |
Gm39149 |
predicted gene, 39149 |
14374 |
0.17 |
| chr5_44922248_44922762 | 2.35 |
4930431F12Rik |
RIKEN cDNA 4930431F12 gene |
43695 |
0.13 |
| chr3_37917085_37917272 | 2.32 |
Gm20755 |
predicted gene, 20755 |
18365 |
0.15 |
| chr4_154346931_154347125 | 2.30 |
Prdm16 |
PR domain containing 16 |
1429 |
0.4 |
| chr5_100568224_100568403 | 2.27 |
Plac8 |
placenta-specific 8 |
3879 |
0.16 |
| chr5_90163210_90163361 | 2.27 |
Eif5al3-ps |
eukaryotic translation initiation factor 5A-like 3, pseudogene |
31056 |
0.18 |
| chr1_162037946_162038745 | 2.26 |
2810442N19Rik |
RIKEN cDNA 2810442N19 gene |
33173 |
0.12 |
| chr13_3709274_3709697 | 2.25 |
Gm47695 |
predicted gene, 47695 |
50340 |
0.09 |
| chr2_24521412_24522064 | 2.24 |
Gm13418 |
predicted gene 13418 |
27670 |
0.11 |
| chr4_28548295_28548623 | 2.23 |
Gm11909 |
predicted gene 11909 |
12247 |
0.19 |
| chr4_143412586_143413277 | 2.23 |
Pramef8 |
PRAME family member 8 |
41 |
0.96 |
| chr2_34983405_34983594 | 2.22 |
Hc |
hemolytic complement |
9837 |
0.12 |
| chr2_146262046_146262668 | 2.20 |
Ralgapa2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
982 |
0.51 |
| chr13_72727296_72727551 | 2.17 |
Gm47890 |
predicted gene, 47890 |
24574 |
0.19 |
| chr9_32720264_32720442 | 2.15 |
Gm27240 |
predicted gene 27240 |
9182 |
0.19 |
| chr15_103032811_103032985 | 2.15 |
Hoxc4 |
homeobox C4 |
1497 |
0.22 |
| chr16_52862236_52862387 | 2.13 |
4930405D01Rik |
RIKEN cDNA 4930405D01 gene |
33181 |
0.2 |
| chr14_16901849_16902015 | 2.12 |
Tpi-rs9 |
triosephosphate isomerase related sequence 9 |
44727 |
0.16 |
| chr3_102197631_102197782 | 2.11 |
Vangl1 |
VANGL planar cell polarity 1 |
199 |
0.92 |
| chr8_115462758_115463065 | 2.09 |
4930488N15Rik |
RIKEN cDNA 4930488N15 gene |
122481 |
0.06 |
| chr9_100271982_100272787 | 2.04 |
Gm28167 |
predicted gene 28167 |
51140 |
0.14 |
| chr2_106487716_106487876 | 2.02 |
Gm14015 |
predicted gene 14015 |
35307 |
0.19 |
| chr13_18363629_18363786 | 2.01 |
Pou6f2 |
POU domain, class 6, transcription factor 2 |
18332 |
0.27 |
| chr3_119946869_119947112 | 2.00 |
Gm18384 |
predicted gene, 18384 |
21741 |
0.24 |
| chr6_65145004_65145200 | 1.99 |
Hpgds |
hematopoietic prostaglandin D synthase |
194 |
0.91 |
| chr10_111923671_111923831 | 1.97 |
Gm47880 |
predicted gene, 47880 |
34737 |
0.12 |
| chr11_12299778_12299982 | 1.97 |
Gm12002 |
predicted gene 12002 |
14634 |
0.26 |
| chr18_69152096_69152441 | 1.97 |
Gm23536 |
predicted gene, 23536 |
21852 |
0.17 |
| chr2_115775579_115775811 | 1.96 |
Meis2 |
Meis homeobox 2 |
93172 |
0.08 |
| chr5_46572933_46573088 | 1.95 |
Gm43093 |
predicted gene 43093 |
70881 |
0.13 |
| chr14_55157316_55157606 | 1.94 |
Gm23596 |
predicted gene, 23596 |
4147 |
0.12 |
| chr5_47752032_47752183 | 1.92 |
Gm3010 |
predicted gene 3010 |
52262 |
0.16 |
| chr8_64791328_64791512 | 1.91 |
Klhl2 |
kelch-like 2, Mayven |
8244 |
0.16 |
| chr6_139777980_139778131 | 1.89 |
Gm37269 |
predicted gene, 37269 |
1625 |
0.4 |
| chr7_144663720_144663871 | 1.89 |
Ano1 |
anoctamin 1, calcium activated chloride channel |
5582 |
0.21 |
| chr6_90650730_90650881 | 1.88 |
Iqsec1 |
IQ motif and Sec7 domain 1 |
11483 |
0.14 |
| chr6_135326951_135327132 | 1.85 |
Gm44275 |
predicted gene, 44275 |
7480 |
0.12 |
| chr6_52161197_52161390 | 1.85 |
Hotairm1 |
Hoxa transcript antisense RNA, myeloid-specific 1 |
2769 |
0.08 |
| chr7_142542880_142543383 | 1.85 |
Mrpl23 |
mitochondrial ribosomal protein L23 |
7027 |
0.1 |
| chr15_18821156_18821696 | 1.85 |
Cdh10 |
cadherin 10 |
1097 |
0.54 |
| chr5_92711815_92711966 | 1.83 |
Gm20500 |
predicted gene 20500 |
24315 |
0.15 |
| chr3_138857779_138858011 | 1.77 |
Tspan5 |
tetraspanin 5 |
2598 |
0.31 |
| chr2_141173180_141173331 | 1.75 |
Zcchc9-ps |
zinc finger, CCHC domain containing 9, pseudogene |
45975 |
0.16 |
| chr17_83103084_83103235 | 1.74 |
Gm46606 |
predicted gene, 46606 |
12476 |
0.19 |
| chr4_95772051_95772219 | 1.73 |
Fggy |
FGGY carbohydrate kinase domain containing |
2626 |
0.39 |
| chr3_130737767_130738023 | 1.73 |
Gm42997 |
predicted gene 42997 |
7294 |
0.12 |
| chr13_50991832_50992018 | 1.71 |
Spin1 |
spindlin 1 |
108955 |
0.06 |
| chr6_128598717_128598972 | 1.71 |
Gm44009 |
predicted gene, 44009 |
10819 |
0.09 |
| chr10_99246954_99247105 | 1.71 |
Gm48884 |
predicted gene, 48884 |
9413 |
0.11 |
| chr9_75590298_75590461 | 1.71 |
Gm48822 |
predicted gene, 48822 |
5270 |
0.15 |
| chr11_94404237_94404528 | 1.71 |
Cacna1g |
calcium channel, voltage-dependent, T type, alpha 1G subunit |
7297 |
0.16 |
| chr6_52177007_52177198 | 1.70 |
5730596B20Rik |
RIKEN cDNA 5730596B20 gene |
396 |
0.56 |
| chr3_120983132_120983283 | 1.70 |
Gm43444 |
predicted gene 43444 |
37569 |
0.16 |
| chr14_9811613_9812129 | 1.69 |
Hmgb3-ps |
Hmgb3 retrotransposed pseudogene |
51231 |
0.15 |
| chr2_124613257_124613683 | 1.69 |
Sema6d |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
2869 |
0.4 |
| chr16_65470607_65470758 | 1.68 |
Gm23659 |
predicted gene, 23659 |
38545 |
0.15 |
| chr6_137126306_137126470 | 1.66 |
Rerg |
RAS-like, estrogen-regulated, growth-inhibitor |
13675 |
0.2 |
| chr10_119024409_119024744 | 1.63 |
Gm47461 |
predicted gene, 47461 |
2906 |
0.23 |
| chr1_42691969_42692512 | 1.63 |
Pantr1 |
POU domain, class 3, transcription factor 3 adjacent noncoding transcript 1 |
853 |
0.48 |
| chr6_8462530_8462681 | 1.62 |
Gm16043 |
predicted gene 16043 |
19150 |
0.14 |
| chr13_94608566_94608905 | 1.62 |
Gm36991 |
predicted gene, 36991 |
10545 |
0.19 |
| chr1_5232314_5232482 | 1.62 |
Gm7182 |
predicted gene 7182 |
44939 |
0.16 |
| chr2_66180561_66180718 | 1.61 |
Gm13618 |
predicted gene 13618 |
7197 |
0.19 |
| chr11_108240891_108241042 | 1.60 |
Gm11655 |
predicted gene 11655 |
59116 |
0.12 |
| chr2_157245722_157245873 | 1.60 |
Mroh8 |
maestro heat-like repeat family member 8 |
12251 |
0.16 |
| chr4_53203595_53203746 | 1.58 |
4930412L05Rik |
RIKEN cDNA 4930412L05 gene |
13891 |
0.18 |
| chr8_82477988_82478139 | 1.58 |
Gm25138 |
predicted gene, 25138 |
68972 |
0.11 |
| chr9_60501613_60501977 | 1.58 |
Thsd4 |
thrombospondin, type I, domain containing 4 |
4669 |
0.17 |
| chr15_25284432_25284672 | 1.58 |
9230109A22Rik |
RIKEN cDNA 9230109A22 gene |
45722 |
0.12 |
| chr7_132645097_132645259 | 1.56 |
Gm15582 |
predicted gene 15582 |
14706 |
0.15 |
| chr4_111452121_111452592 | 1.56 |
Bend5 |
BEN domain containing 5 |
19037 |
0.21 |
| chr9_54782563_54782714 | 1.54 |
Crabp1 |
cellular retinoic acid binding protein I |
17890 |
0.15 |
| chr1_22538228_22538420 | 1.53 |
Rims1 |
regulating synaptic membrane exocytosis 1 |
25795 |
0.27 |
| chr16_25584812_25584969 | 1.52 |
Trp63 |
transformation related protein 63 |
98873 |
0.08 |
| chr2_65568035_65568186 | 1.52 |
Scn3a |
sodium channel, voltage-gated, type III, alpha |
483 |
0.85 |
| chr4_81744878_81745052 | 1.52 |
Gm11412 |
predicted gene 11412 |
1819 |
0.45 |
| chr13_102624971_102625122 | 1.52 |
Gm47014 |
predicted gene, 47014 |
20513 |
0.2 |
| chr3_123386813_123386972 | 1.51 |
Mettl14 |
methyltransferase like 14 |
784 |
0.56 |
| chr11_45922045_45922196 | 1.50 |
Lsm11 |
U7 snRNP-specific Sm-like protein LSM11 |
22815 |
0.13 |
| chr14_12026153_12026339 | 1.50 |
Gm3848 |
predicted gene 3848 |
45023 |
0.15 |
| chr10_92076795_92077161 | 1.50 |
Rmst |
rhabdomyosarcoma 2 associated transcript (non-coding RNA) |
1349 |
0.46 |
| chr18_80984858_80985066 | 1.48 |
Sall3 |
spalt like transcription factor 3 |
1574 |
0.28 |
| chr5_37893454_37893628 | 1.48 |
Gm20052 |
predicted gene, 20052 |
231 |
0.94 |
| chr4_95278814_95279018 | 1.48 |
Gm12708 |
predicted gene 12708 |
85554 |
0.08 |
| chr6_86775025_86775336 | 1.46 |
Anxa4 |
annexin A4 |
9270 |
0.14 |
| chr5_16045560_16045728 | 1.46 |
Cacna2d1 |
calcium channel, voltage-dependent, alpha2/delta subunit 1 |
19930 |
0.19 |
| chr2_138708817_138708988 | 1.46 |
Mir1952 |
microRNA 1952 |
111027 |
0.07 |
| chr15_6025556_6025892 | 1.44 |
Gm8047 |
predicted gene 8047 |
9237 |
0.26 |
| chr16_57484144_57484357 | 1.44 |
Filip1l |
filamin A interacting protein 1-like |
64992 |
0.11 |
| chr1_138965657_138965808 | 1.43 |
Dennd1b |
DENN/MADD domain containing 1B |
1863 |
0.18 |
| chr15_27259054_27259330 | 1.43 |
Gm19111 |
predicted gene, 19111 |
108747 |
0.07 |
| chr16_36817082_36817234 | 1.43 |
Eaf2 |
ELL associated factor 2 |
8453 |
0.11 |
| chr4_139639864_139640099 | 1.42 |
Mir7020 |
microRNA 7020 |
4061 |
0.17 |
| chr9_23041547_23042363 | 1.42 |
Bmper |
BMP-binding endothelial regulator |
181121 |
0.03 |
| chr1_82766345_82766504 | 1.41 |
Tm4sf20 |
transmembrane 4 L six family member 20 |
2037 |
0.17 |
| chr13_25220533_25220766 | 1.40 |
Dcdc2a |
doublecortin domain containing 2a |
33195 |
0.17 |
| chr1_192735838_192736339 | 1.39 |
Hhat |
hedgehog acyltransferase |
1192 |
0.48 |
| chr18_43687992_43688219 | 1.39 |
Jakmip2 |
janus kinase and microtubule interacting protein 2 |
332 |
0.9 |
| chr13_8526132_8526287 | 1.39 |
Gm48262 |
predicted gene, 48262 |
26811 |
0.23 |
| chr15_103135415_103135966 | 1.38 |
Gm49477 |
predicted gene, 49477 |
191 |
0.89 |
| chr18_46213235_46213386 | 1.38 |
Trim36 |
tripartite motif-containing 36 |
703 |
0.66 |
| chr10_122910780_122911062 | 1.37 |
Ppm1h |
protein phosphatase 1H (PP2C domain containing) |
15553 |
0.2 |
| chrX_49288700_49288851 | 1.37 |
Enox2 |
ecto-NOX disulfide-thiol exchanger 2 |
516 |
0.84 |
| chr15_25647400_25647551 | 1.37 |
Gm48996 |
predicted gene, 48996 |
12848 |
0.18 |
| chr8_61451835_61452034 | 1.36 |
Cbr4 |
carbonyl reductase 4 |
35800 |
0.16 |
| chr4_14273616_14273932 | 1.36 |
Gm24908 |
predicted gene, 24908 |
34156 |
0.18 |
| chr14_105920859_105921276 | 1.36 |
Spry2 |
sprouty RTK signaling antagonist 2 |
24248 |
0.21 |
| chr3_10336279_10336457 | 1.35 |
Gm38303 |
predicted gene, 38303 |
687 |
0.36 |
| chr1_83117273_83117561 | 1.35 |
Ccl20 |
chemokine (C-C motif) ligand 20 |
624 |
0.62 |
| chr1_172222161_172222312 | 1.35 |
Casq1 |
calsequestrin 1 |
2368 |
0.15 |
| chr2_181653165_181653343 | 1.34 |
Gm29797 |
predicted gene, 29797 |
3780 |
0.11 |
| chr4_148389759_148389933 | 1.34 |
Gm13200 |
predicted gene 13200 |
30386 |
0.12 |
| chr2_164518601_164518762 | 1.34 |
Pigt |
phosphatidylinositol glycan anchor biosynthesis, class T |
18765 |
0.08 |
| chr7_19156164_19156362 | 1.34 |
Snrpd2 |
small nuclear ribonucleoprotein D2 |
6404 |
0.08 |
| chr8_80204151_80204327 | 1.34 |
Gm45430 |
predicted gene 45430 |
9040 |
0.25 |
| chr14_102031439_102031595 | 1.33 |
Gm5854 |
predicted gene 5854 |
48754 |
0.18 |
| chr6_136761643_136761794 | 1.33 |
Gucy2c |
guanylate cyclase 2c |
20024 |
0.11 |
| chr13_21160100_21160442 | 1.32 |
Olfr1368 |
olfactory receptor 1368 |
14404 |
0.12 |
| chr7_65955319_65955578 | 1.32 |
Pcsk6 |
proprotein convertase subtilisin/kexin type 6 |
2436 |
0.32 |
| chr3_117873618_117873778 | 1.31 |
Snx7 |
sorting nexin 7 |
4762 |
0.22 |
| chr7_45917070_45917525 | 1.30 |
Tmem143 |
transmembrane protein 143 |
1299 |
0.21 |
| chr8_26770434_26770629 | 1.29 |
Gm45647 |
predicted gene 45647 |
9784 |
0.17 |
| chr12_37541109_37541260 | 1.29 |
Gm48660 |
predicted gene, 48660 |
28095 |
0.22 |
| chr3_60097928_60098098 | 1.29 |
Sucnr1 |
succinate receptor 1 |
16111 |
0.18 |
| chr14_7824721_7825158 | 1.29 |
Flnb |
filamin, beta |
6982 |
0.16 |
| chr10_105806591_105806900 | 1.28 |
Gm48298 |
predicted gene, 48298 |
2122 |
0.26 |
| chr10_96545457_96545608 | 1.28 |
Gm48505 |
predicted gene, 48505 |
21550 |
0.18 |
| chr2_44304639_44304868 | 1.28 |
Gm22867 |
predicted gene, 22867 |
124182 |
0.06 |
| chr15_42373080_42373231 | 1.27 |
Gm49453 |
predicted gene, 49453 |
26600 |
0.18 |
| chr13_60497474_60497625 | 1.27 |
A530001N23Rik |
RIKEN cDNA A530001N23 gene |
8741 |
0.17 |
| chr7_58736928_58737203 | 1.26 |
Gm44937 |
predicted gene 44937 |
16866 |
0.18 |
| chrX_95658622_95659031 | 1.26 |
Zc4h2 |
zinc finger, C4H2 domain containing |
317 |
0.9 |
| chr5_51144930_51145171 | 1.25 |
Gm44377 |
predicted gene, 44377 |
78624 |
0.1 |
| chr16_55611251_55611402 | 1.25 |
Gm23003 |
predicted gene, 23003 |
16438 |
0.25 |
| chr14_98084865_98085016 | 1.25 |
Gm16331 |
predicted gene 16331 |
33569 |
0.21 |
| chr11_18749927_18750078 | 1.25 |
Gm28401 |
predicted gene 28401 |
6736 |
0.18 |
| chr3_144849567_144849863 | 1.25 |
Clca3b |
chloride channel accessory 3B |
358 |
0.79 |
| chr14_96218177_96218332 | 1.25 |
Gm15515 |
predicted gene 15515 |
13699 |
0.3 |
| chr15_16728489_16728641 | 1.25 |
Cdh9 |
cadherin 9 |
191 |
0.97 |
| chr2_133399061_133399369 | 1.25 |
A430048G15Rik |
RIKEN cDNA A430048G15 gene |
32880 |
0.18 |
| chrY_90793215_90793563 | 1.25 |
Gm47283 |
predicted gene, 47283 |
2938 |
0.25 |
| chr7_84237857_84238008 | 1.24 |
Gm44827 |
predicted gene 44827 |
5013 |
0.17 |
| chr6_21863731_21863882 | 1.24 |
Gm3289 |
predicted gene 3289 |
5628 |
0.2 |
| chr11_32002527_32003111 | 1.24 |
Nsg2 |
neuron specific gene family member 2 |
2317 |
0.33 |
| chr9_75960650_75960801 | 1.24 |
Gm34005 |
predicted gene, 34005 |
3479 |
0.2 |
| chr7_3411029_3411836 | 1.23 |
Gm23450 |
predicted gene, 23450 |
3634 |
0.11 |
| chr8_55115169_55115732 | 1.23 |
Gm8734 |
predicted gene 8734 |
33735 |
0.14 |
| chr8_91613624_91613901 | 1.23 |
Gm3235 |
predicted gene 3235 |
15967 |
0.13 |
| chr5_70842284_70842796 | 1.23 |
Gabrg1 |
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 1 |
77 |
0.99 |
| chr4_89177615_89177779 | 1.23 |
Mtap |
methylthioadenosine phosphorylase |
1323 |
0.34 |
| chr9_77160407_77160574 | 1.22 |
Mlip |
muscular LMNA-interacting protein |
21788 |
0.2 |
| chr16_28841061_28841259 | 1.22 |
Mb21d2 |
Mab-21 domain containing 2 |
4793 |
0.32 |
| chr6_142701662_142702025 | 1.22 |
Abcc9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
375 |
0.88 |
| chr2_106149818_106150014 | 1.21 |
Dcdc5 |
doublecortin domain containing 5 |
72141 |
0.1 |
| chr10_99037188_99037357 | 1.20 |
Gm48761 |
predicted gene, 48761 |
2200 |
0.29 |
| chr4_83656810_83657017 | 1.20 |
Ccdc171 |
coiled-coil domain containing 171 |
39042 |
0.14 |
| chr13_44026713_44026864 | 1.20 |
Gm33489 |
predicted gene, 33489 |
8266 |
0.22 |
| chr3_156560293_156560473 | 1.20 |
4930570G19Rik |
RIKEN cDNA 4930570G19 gene |
1199 |
0.41 |
| chr5_125264626_125264993 | 1.19 |
Gm32585 |
predicted gene, 32585 |
8006 |
0.18 |
| chr4_129237582_129237740 | 1.19 |
C77080 |
expressed sequence C77080 |
1513 |
0.28 |
| chr4_72627577_72627857 | 1.19 |
Gm11235 |
predicted gene 11235 |
85051 |
0.1 |
| chr15_50888825_50889690 | 1.18 |
Mir1907 |
microRNA 1907 |
143 |
0.59 |
| chr17_59383142_59383349 | 1.18 |
Gm23769 |
predicted gene, 23769 |
157402 |
0.04 |
| chr4_54945565_54946178 | 1.18 |
Zfp462 |
zinc finger protein 462 |
823 |
0.73 |
| chr11_38856747_38856914 | 1.17 |
Gm23520 |
predicted gene, 23520 |
58553 |
0.17 |
| chr14_24550971_24551440 | 1.17 |
Rps24 |
ribosomal protein S24 |
60056 |
0.09 |
| chr18_40414592_40414743 | 1.17 |
Kctd16 |
potassium channel tetramerisation domain containing 16 |
6582 |
0.23 |
| chr2_65620615_65620890 | 1.17 |
Scn2a |
sodium channel, voltage-gated, type II, alpha |
19 |
0.98 |
| chr8_92460573_92460724 | 1.17 |
Gm45336 |
predicted gene 45336 |
5119 |
0.28 |
| chr6_126553960_126554111 | 1.17 |
Gm33320 |
predicted gene, 33320 |
881 |
0.53 |
| chr12_7611773_7612006 | 1.16 |
Gm9139 |
predicted gene 9139 |
193765 |
0.03 |
| chr16_56721143_56721294 | 1.15 |
Tfg |
Trk-fused gene |
3768 |
0.26 |
| chr18_31950013_31950364 | 1.15 |
Gpr17 |
G protein-coupled receptor 17 |
552 |
0.68 |
| chr10_125809855_125810006 | 1.15 |
Lrig3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
156238 |
0.04 |
| chr5_81020036_81020210 | 1.15 |
Adgrl3 |
adhesion G protein-coupled receptor L3 |
15 |
0.99 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.5 | 1.8 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.4 | 1.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.3 | 1.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.3 | 1.0 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.3 | 0.9 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 1.2 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.3 | 0.8 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 0.5 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.2 | 0.9 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.2 | 0.4 | GO:2000302 | positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.2 | 1.2 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.2 | 0.2 | GO:0072069 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.2 | 0.6 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.2 | 0.7 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) |
| 0.2 | 0.9 | GO:0032730 | positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.2 | 0.7 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.2 | 0.5 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.2 | 0.8 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.2 | 0.8 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.2 | 0.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.2 | 0.8 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 0.5 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 1.0 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.4 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.8 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.3 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 | 1.0 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 1.4 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.4 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.1 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.3 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.3 | GO:1903626 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.1 | 0.3 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.1 | 0.3 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.9 | GO:0060013 | righting reflex(GO:0060013) |
| 0.1 | 0.2 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.1 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 0.1 | 0.3 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.1 | 0.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.4 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.1 | 0.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.1 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.7 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.7 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.7 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.6 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.5 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.3 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 0.2 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.3 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.1 | 0.3 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 1.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.1 | 0.2 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.4 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.1 | 0.2 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.1 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.3 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.1 | 0.2 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.1 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.1 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.1 | GO:0007442 | hindgut morphogenesis(GO:0007442) |
| 0.1 | 0.8 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.1 | GO:2000449 | regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000449) |
| 0.1 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.1 | 1.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.2 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.1 | 0.1 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.4 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.2 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 0.2 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.3 | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis(GO:0061309) |
| 0.1 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.9 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.1 | 0.3 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.2 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.4 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.2 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0072178 | nephric duct development(GO:0072176) nephric duct morphogenesis(GO:0072178) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.2 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.0 | 0.2 | GO:0044597 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.3 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.0 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.5 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.0 | GO:0072008 | mesangial cell differentiation(GO:0072007) glomerular mesangial cell differentiation(GO:0072008) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.0 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.2 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.6 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.2 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.0 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 2.9 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.2 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.2 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.0 | 0.1 | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 0.0 | GO:0060847 | endothelial cell fate specification(GO:0060847) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.0 | 0.6 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.6 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.2 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.0 | GO:0097475 | motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.1 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.2 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.0 | 0.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 0.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0060347 | heart trabecula formation(GO:0060347) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.0 | 0.1 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.1 | GO:0010991 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.1 | GO:0072338 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.6 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0010912 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.0 | 2.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.0 | 0.2 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.1 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.0 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.5 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.2 | GO:0010388 | cullin deneddylation(GO:0010388) |
| 0.0 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:1901525 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.3 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.2 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.0 | GO:0061218 | negative regulation of mesonephros development(GO:0061218) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.0 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.0 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.0 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.0 | 0.0 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.0 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.8 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.3 | GO:0033273 | response to vitamin(GO:0033273) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.0 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.0 | 0.1 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0072044 | collecting duct development(GO:0072044) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.0 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.0 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.0 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.0 | GO:0072174 | metanephric tubule formation(GO:0072174) |
| 0.0 | 0.0 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.5 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.0 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:0060374 | mast cell differentiation(GO:0060374) regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.0 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.0 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.0 | GO:0002692 | negative regulation of cellular extravasation(GO:0002692) |
| 0.0 | 0.0 | GO:0031054 | pre-miRNA processing(GO:0031054) |
| 0.0 | 0.0 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.0 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:1900044 | regulation of protein K63-linked ubiquitination(GO:1900044) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0036445 | neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.2 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.0 | GO:0072198 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0045779 | negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.1 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.0 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.0 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0061050 | regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.0 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.0 | 0.1 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.0 | GO:0009196 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.0 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.0 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.0 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.0 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.0 | 0.0 | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035358) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.0 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.0 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.0 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.0 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.0 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.0 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 0.9 | GO:0036396 | MIS complex(GO:0036396) |
| 0.2 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.8 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.3 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.9 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.9 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0034679 | integrin alpha9-beta1 complex(GO:0034679) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.0 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.0 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.7 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.0 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.0 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.0 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.0 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.3 | 1.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.2 | 0.9 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 0.8 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 0.5 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 0.7 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.9 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.8 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 0.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 1.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.1 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.6 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.2 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 1.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.2 | GO:0001032 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.1 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.4 | GO:0035410 | dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.0 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.4 | GO:0034848 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.3 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 1.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0008932 | lytic endotransglycosylase activity(GO:0008932) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0019107 | myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.5 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.0 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.0 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.0 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.0 | 0.1 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0032357 | oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0052828 | JUN kinase phosphatase activity(GO:0008579) phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) inositol-4,5-bisphosphate 5-phosphatase activity(GO:0030487) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.1 | GO:0034548 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.0 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 1.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.0 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.0 | REACTOME ORC1 REMOVAL FROM CHROMATIN | Genes involved in Orc1 removal from chromatin |
| 0.0 | 0.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |