Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
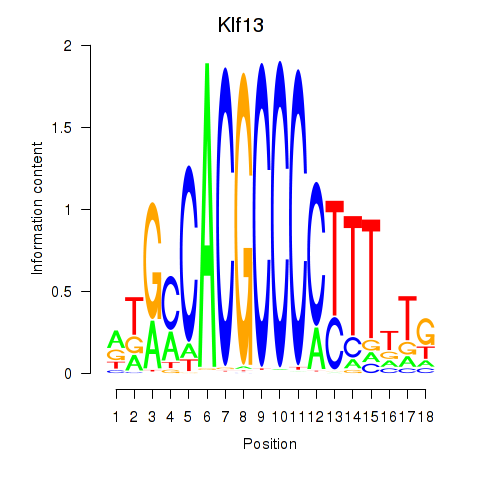
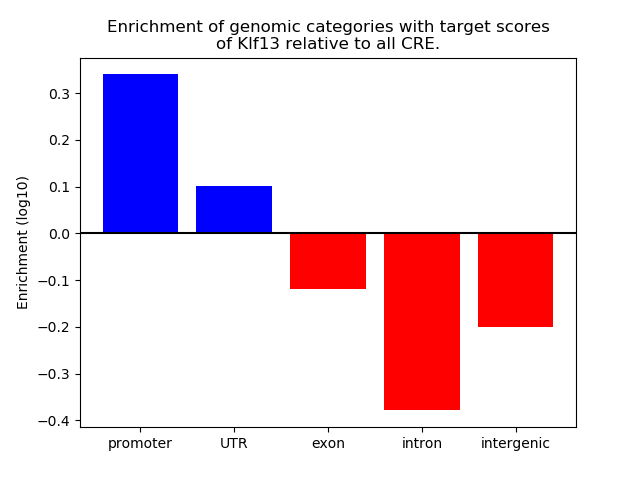
Results for Klf13
Z-value: 0.96
Transcription factors associated with Klf13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Klf13
|
ENSMUSG00000052040.9 | Kruppel-like factor 13 |
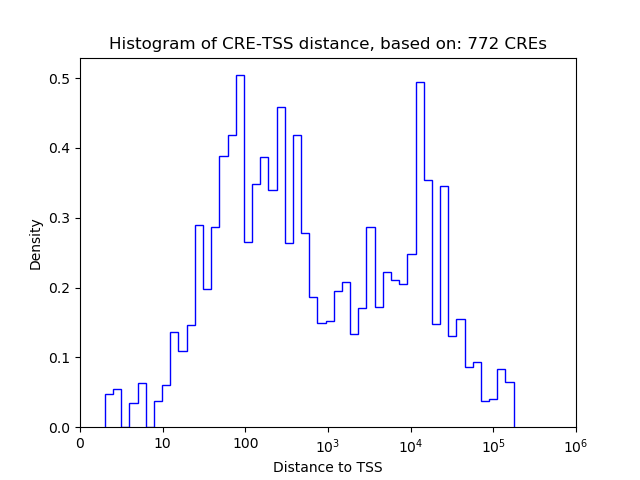
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_63937176_63937806 | Klf13 | 1159 | 0.384575 | -0.61 | 5.9e-07 | Click! |
| chr7_63938917_63939336 | Klf13 | 211 | 0.908055 | -0.58 | 4.2e-06 | Click! |
| chr7_63939410_63939940 | Klf13 | 760 | 0.551739 | -0.58 | 4.2e-06 | Click! |
| chr7_63937825_63938018 | Klf13 | 994 | 0.442186 | -0.55 | 1.4e-05 | Click! |
| chr7_63941140_63941323 | Klf13 | 2316 | 0.209409 | -0.54 | 2.1e-05 | Click! |
Activity of the Klf13 motif across conditions
Conditions sorted by the z-value of the Klf13 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
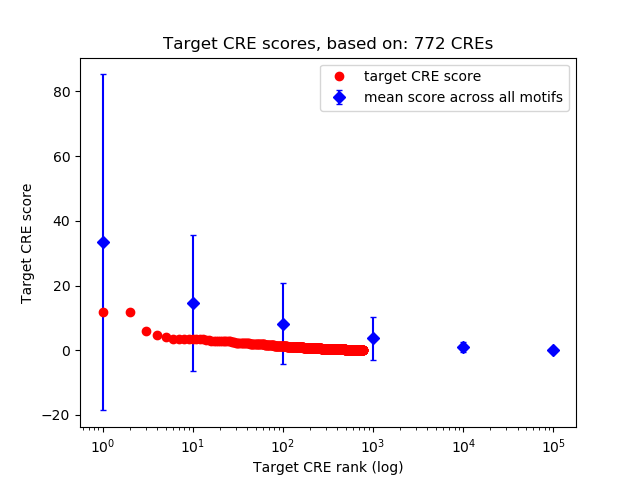
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_124439906_124440949 | 11.92 |
Ppp2r3d |
protein phosphatase 2 (formerly 2A), regulatory subunit B'', delta |
441 |
0.79 |
| chrY_90739614_90740540 | 11.85 |
Mid1-ps1 |
midline 1, pseudogene 1 |
12980 |
0.18 |
| chr8_70476397_70477967 | 5.86 |
Klhl26 |
kelch-like 26 |
214 |
0.86 |
| chr13_112440041_112440301 | 4.57 |
Gm48879 |
predicted gene, 48879 |
3575 |
0.18 |
| chr10_81106448_81106795 | 4.21 |
Map2k2 |
mitogen-activated protein kinase kinase 2 |
276 |
0.75 |
| chr9_18473066_18474201 | 3.53 |
Zfp558 |
zinc finger protein 558 |
74 |
0.95 |
| chr5_9726061_9726270 | 3.48 |
Grm3 |
glutamate receptor, metabotropic 3 |
995 |
0.6 |
| chr10_22191194_22191366 | 3.45 |
Raet1e |
retinoic acid early transcript 1E |
17361 |
0.1 |
| chr14_120388810_120389107 | 3.43 |
Gm26679 |
predicted gene, 26679 |
27 |
0.98 |
| chr12_3236518_3237725 | 3.42 |
Rab10os |
RAB10, member RAS oncogene family, opposite strand |
510 |
0.74 |
| chr11_42314956_42315148 | 3.41 |
Gabra6 |
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 6 |
5997 |
0.32 |
| chr17_56626470_56627486 | 3.37 |
Lonp1 |
lon peptidase 1, mitochondrial |
91 |
0.92 |
| chr10_22381380_22381531 | 3.33 |
Raet1d |
retinoic acid early transcript delta |
19561 |
0.12 |
| chr2_57102640_57102858 | 3.27 |
Nr4a2 |
nuclear receptor subfamily 4, group A, member 2 |
10323 |
0.2 |
| chr12_108333504_108334768 | 3.12 |
Cyp46a1 |
cytochrome P450, family 46, subfamily a, polypeptide 1 |
245 |
0.91 |
| chr13_60737684_60738259 | 2.96 |
Dapk1 |
death associated protein kinase 1 |
16025 |
0.15 |
| chr15_82293240_82293859 | 2.94 |
Wbp2nl |
WBP2 N-terminal like |
5405 |
0.09 |
| chr17_56664334_56664784 | 2.91 |
Ranbp3 |
RAN binding protein 3 |
8735 |
0.11 |
| chr12_102468744_102469733 | 2.87 |
Golga5 |
golgi autoantigen, golgin subfamily a, 5 |
46 |
0.97 |
| chr9_119119523_119119674 | 2.85 |
Dlec1 |
deleted in lung and esophageal cancer 1 |
56 |
0.96 |
| chr1_5588268_5588565 | 2.84 |
Oprk1 |
opioid receptor, kappa 1 |
50 |
0.98 |
| chr4_128884076_128884722 | 2.84 |
Trim62 |
tripartite motif-containing 62 |
811 |
0.57 |
| chr8_84990346_84991110 | 2.83 |
Hook2 |
hook microtubule tethering protein 2 |
86 |
0.9 |
| chr5_109552541_109552889 | 2.80 |
Gm8493 |
predicted gene 8493 |
1441 |
0.33 |
| chr14_14347096_14348750 | 2.79 |
Gm48860 |
predicted gene, 48860 |
659 |
0.44 |
| chr4_152886919_152887080 | 2.72 |
Gm25779 |
predicted gene, 25779 |
126585 |
0.05 |
| chr5_146384860_146385320 | 2.68 |
Wasf3 |
WAS protein family, member 3 |
105 |
0.97 |
| chr5_73599424_73599575 | 2.58 |
Lrrc66 |
leucine rich repeat containing 66 |
30813 |
0.13 |
| chr13_95994774_95995004 | 2.57 |
Sv2c |
synaptic vesicle glycoprotein 2c |
4663 |
0.25 |
| chr1_157798948_157799099 | 2.34 |
Gm38256 |
predicted gene, 38256 |
141362 |
0.04 |
| chr7_49496969_49497490 | 2.26 |
Gm38059 |
predicted gene, 38059 |
27517 |
0.2 |
| chr8_120509117_120509877 | 2.24 |
Gm26971 |
predicted gene, 26971 |
11118 |
0.13 |
| chr7_5019965_5020617 | 2.22 |
Zfp865 |
zinc finger protein 865 |
85 |
0.7 |
| chr10_80741710_80742767 | 2.20 |
Ap3d1 |
adaptor-related protein complex 3, delta 1 subunit |
26 |
0.95 |
| chr8_69832079_69833191 | 2.15 |
Pbx4 |
pre B cell leukemia homeobox 4 |
2 |
0.96 |
| chr18_35965723_35965937 | 2.15 |
Psd2 |
pleckstrin and Sec7 domain containing 2 |
725 |
0.59 |
| chr11_44617130_44617322 | 2.14 |
Ebf1 |
early B cell factor 1 |
91 |
0.67 |
| chr6_56568728_56569302 | 2.12 |
Pde1c |
phosphodiesterase 1C |
88 |
0.97 |
| chr8_70905797_70906132 | 2.11 |
Map1s |
microtubule-associated protein 1S |
18 |
0.94 |
| chr7_29210966_29212124 | 2.09 |
Catsperg1 |
cation channel sperm associated auxiliary subunit gamma 1 |
50 |
0.94 |
| chr7_135863781_135863970 | 2.08 |
Gm45241 |
predicted gene 45241 |
9427 |
0.16 |
| chr6_85068662_85069731 | 2.08 |
Exoc6b |
exocyst complex component 6B |
261 |
0.84 |
| chr3_132855591_132855776 | 2.07 |
Gm9403 |
predicted gene 9403 |
13287 |
0.14 |
| chr19_5447521_5447969 | 2.06 |
Fosl1 |
fos-like antigen 1 |
40 |
0.9 |
| chr15_37792454_37792630 | 2.04 |
Ncald |
neurocalcin delta |
28 |
0.98 |
| chr9_114528016_114528485 | 2.02 |
C130032M10Rik |
RIKEN cDNA C130032M10 gene |
12553 |
0.16 |
| chr2_145674795_145675792 | 2.00 |
Rin2 |
Ras and Rab interactor 2 |
74 |
0.98 |
| chr4_154948181_154948553 | 2.00 |
Hes5 |
hes family bHLH transcription factor 5 |
12556 |
0.11 |
| chr2_32875948_32876955 | 1.95 |
Fam129b |
family with sequence similarity 129, member B |
337 |
0.54 |
| chr2_72242138_72242289 | 1.95 |
Rapgef4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
81 |
0.97 |
| chr5_38560766_38562198 | 1.94 |
Wdr1 |
WD repeat domain 1 |
61 |
0.97 |
| chr2_26444885_26445708 | 1.92 |
Sec16a |
SEC16 homolog A, endoplasmic reticulum export factor |
80 |
0.82 |
| chr17_13668639_13669018 | 1.91 |
2700054A10Rik |
RIKEN cDNA 2700054A10 gene |
63 |
0.97 |
| chr4_120854573_120855281 | 1.89 |
Rims3 |
regulating synaptic membrane exocytosis 3 |
108 |
0.95 |
| chr11_67081283_67081678 | 1.89 |
Myh3 |
myosin, heavy polypeptide 3, skeletal muscle, embryonic |
3180 |
0.17 |
| chrX_6778341_6778533 | 1.88 |
Dgkk |
diacylglycerol kinase kappa |
869 |
0.74 |
| chr16_90400411_90400594 | 1.84 |
Hunk |
hormonally upregulated Neu-associated kinase |
42 |
0.97 |
| chr7_19118025_19118835 | 1.83 |
Gm4969 |
predicted gene 4969 |
62 |
0.92 |
| chr1_172617765_172617949 | 1.82 |
Dusp23 |
dual specificity phosphatase 23 |
15105 |
0.12 |
| chr8_46470414_46471778 | 1.81 |
Acsl1 |
acyl-CoA synthetase long-chain family member 1 |
24 |
0.97 |
| chr11_70026722_70027238 | 1.80 |
Dlg4 |
discs large MAGUK scaffold protein 4 |
19 |
0.93 |
| chr7_64569803_64569954 | 1.78 |
Gm44721 |
predicted gene 44721 |
40453 |
0.15 |
| chr8_71727942_71728505 | 1.74 |
Fcho1 |
FCH domain only 1 |
2507 |
0.15 |
| chr7_18950998_18951210 | 1.69 |
Nova2 |
NOVA alternative splicing regulator 2 |
25216 |
0.07 |
| chr4_119733256_119734302 | 1.64 |
Hivep3 |
human immunodeficiency virus type I enhancer binding protein 3 |
5 |
0.98 |
| chr5_41764546_41764740 | 1.64 |
Nkx3-2 |
NK3 homeobox 2 |
142 |
0.97 |
| chr5_144357735_144358915 | 1.64 |
Dmrt1i |
Dmrt1 interacting ncRNA |
200 |
0.51 |
| chr17_11664742_11664947 | 1.62 |
Gm10513 |
predicted gene 10513 |
67501 |
0.13 |
| chr8_71628994_71630252 | 1.60 |
Mir6769b |
microRNA 6769b |
1483 |
0.21 |
| chr8_12873206_12874084 | 1.59 |
Mcf2l |
mcf.2 transforming sequence-like |
161 |
0.92 |
| chr14_61172444_61173564 | 1.59 |
Sacs |
sacsin |
14 |
0.98 |
| chr9_91228155_91228326 | 1.56 |
Gm29602 |
predicted gene 29602 |
12225 |
0.18 |
| chr14_67308172_67308323 | 1.55 |
Gm6878 |
predicted gene 6878 |
6464 |
0.18 |
| chr5_101665294_101665572 | 1.55 |
Nkx6-1 |
NK6 homeobox 1 |
437 |
0.83 |
| chr7_78990295_78990597 | 1.54 |
Gm26633 |
predicted gene, 26633 |
62877 |
0.08 |
| chr18_84685090_84685924 | 1.48 |
Cndp2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
120 |
0.94 |
| chr11_47377484_47377672 | 1.48 |
Sgcd |
sarcoglycan, delta (dystrophin-associated glycoprotein) |
1944 |
0.5 |
| chr3_88536213_88537501 | 1.46 |
Mir1905 |
microRNA 1905 |
475 |
0.58 |
| chr8_112569720_112569921 | 1.46 |
Cntnap4 |
contactin associated protein-like 4 |
223 |
0.52 |
| chr2_33543695_33544688 | 1.45 |
Gm13530 |
predicted gene 13530 |
54409 |
0.1 |
| chr13_55012325_55012476 | 1.44 |
Hk3 |
hexokinase 3 |
8504 |
0.15 |
| chr5_43289896_43290047 | 1.42 |
6030400A10Rik |
RIKEN cDNA 6030400A10 gene |
24814 |
0.14 |
| chr16_22919964_22920479 | 1.41 |
Fetub |
fetuin beta |
16 |
0.96 |
| chr3_34642307_34643037 | 1.39 |
Gm42692 |
predicted gene 42692 |
592 |
0.6 |
| chr10_93833649_93834340 | 1.38 |
Usp44 |
ubiquitin specific peptidase 44 |
176 |
0.93 |
| chr5_139790880_139791640 | 1.38 |
Mafk |
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) |
253 |
0.87 |
| chr19_29325028_29325544 | 1.36 |
Insl6 |
insulin-like 6 |
70 |
0.96 |
| chr14_54958021_54958172 | 1.35 |
Myh6 |
myosin, heavy polypeptide 6, cardiac muscle, alpha |
189 |
0.82 |
| chr3_39220025_39220213 | 1.33 |
Gm43008 |
predicted gene 43008 |
2273 |
0.42 |
| chr5_142701305_142702538 | 1.32 |
Slc29a4 |
solute carrier family 29 (nucleoside transporters), member 4 |
180 |
0.95 |
| chrX_141725754_141725924 | 1.32 |
Irs4 |
insulin receptor substrate 4 |
576 |
0.59 |
| chr19_36409278_36410322 | 1.32 |
Pcgf5 |
polycomb group ring finger 5 |
52 |
0.97 |
| chr6_91409483_91409682 | 1.31 |
Wnt7a |
wingless-type MMTV integration site family, member 7A |
1781 |
0.2 |
| chr2_173658978_173659962 | 1.30 |
Ppp4r1l-ps |
protein phosphatase 4, regulatory subunit 1-like, pseudogene |
34 |
0.8 |
| chr11_120349471_120350657 | 1.30 |
0610009L18Rik |
RIKEN cDNA 0610009L18 gene |
1386 |
0.18 |
| chr11_117425997_117426148 | 1.29 |
Gm11732 |
predicted gene 11732 |
106 |
0.95 |
| chr7_16129395_16130325 | 1.25 |
Slc8a2 |
solute carrier family 8 (sodium/calcium exchanger), member 2 |
34 |
0.96 |
| chr7_18963587_18963738 | 1.24 |
Nanos2 |
nanos C2HC-type zinc finger 2 |
23738 |
0.07 |
| chr6_22874878_22875647 | 1.24 |
Ptprz1 |
protein tyrosine phosphatase, receptor type Z, polypeptide 1 |
240 |
0.94 |
| chr5_117125754_117126245 | 1.23 |
Taok3 |
TAO kinase 3 |
302 |
0.86 |
| chr14_70443361_70444026 | 1.22 |
Mir320 |
microRNA 320 |
183 |
0.56 |
| chr8_64946989_64947925 | 1.22 |
Tmem192 |
transmembrane protein 192 |
203 |
0.55 |
| chr12_117447218_117447625 | 1.22 |
Rapgef5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
69058 |
0.12 |
| chr16_34004064_34004278 | 1.22 |
Gm49594 |
predicted gene, 49594 |
3286 |
0.22 |
| chr3_103102012_103103284 | 1.18 |
Dennd2c |
DENN/MADD domain containing 2C |
12 |
0.96 |
| chr13_13783865_13785053 | 1.18 |
Gng4 |
guanine nucleotide binding protein (G protein), gamma 4 |
130 |
0.96 |
| chr8_71374903_71376449 | 1.18 |
Nr2f6 |
nuclear receptor subfamily 2, group F, member 6 |
114 |
0.92 |
| chr13_55474642_55474940 | 1.16 |
Mir6944 |
microRNA 6944 |
2985 |
0.11 |
| chr12_108004757_108004945 | 1.15 |
Bcl11b |
B cell leukemia/lymphoma 11B |
1249 |
0.59 |
| chr8_4677704_4678740 | 1.15 |
Gm7461 |
predicted gene 7461 |
143 |
0.62 |
| chr16_77645068_77645328 | 1.15 |
Mir125b-2 |
microRNA 125b-2 |
1075 |
0.28 |
| chr15_98951811_98951974 | 1.14 |
Gm49450 |
predicted gene, 49450 |
1659 |
0.16 |
| chr4_86748363_86749478 | 1.14 |
Dennd4c |
DENN/MADD domain containing 4C |
365 |
0.89 |
| chr11_33512825_33513920 | 1.14 |
Ranbp17 |
RAN binding protein 17 |
258 |
0.93 |
| chr2_135536302_135536795 | 1.12 |
9630028H03Rik |
RIKEN cDNA 9630028H03 gene |
46748 |
0.17 |
| chr7_37820786_37820956 | 1.11 |
Gm22302 |
predicted gene, 22302 |
3476 |
0.3 |
| chr7_27178071_27179218 | 1.10 |
Rab4b |
RAB4B, member RAS oncogene family |
166 |
0.84 |
| chr2_152707269_152707927 | 1.10 |
H13 |
histocompatibility 13 |
15759 |
0.09 |
| chr13_99444919_99445223 | 1.09 |
Map1b |
microtubule-associated protein 1B |
604 |
0.74 |
| chr11_35838430_35838735 | 1.09 |
Rars |
arginyl-tRNA synthetase |
4076 |
0.2 |
| chr1_132559167_132559406 | 1.09 |
Cntn2 |
contactin 2 |
16030 |
0.16 |
| chr6_116673486_116674013 | 1.09 |
Rassf4 |
Ras association (RalGDS/AF-6) domain family member 4 |
22 |
0.96 |
| chr13_12317316_12317467 | 1.08 |
Actn2 |
actinin alpha 2 |
23333 |
0.15 |
| chr14_99045440_99046902 | 1.07 |
Mzt1 |
mitotic spindle organizing protein 1 |
37 |
0.51 |
| chr2_73590227_73590410 | 1.06 |
Chrna1os |
cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle), opposite strand |
5458 |
0.15 |
| chr13_39001535_39001686 | 1.06 |
Slc35b3 |
solute carrier family 35, member B3 |
40735 |
0.15 |
| chr8_71686115_71686959 | 1.06 |
Insl3 |
insulin-like 3 |
2677 |
0.12 |
| chr2_135324013_135324227 | 1.05 |
Plcb1 |
phospholipase C, beta 1 |
74947 |
0.12 |
| chr6_108828087_108829378 | 1.05 |
Edem1 |
ER degradation enhancer, mannosidase alpha-like 1 |
58 |
0.98 |
| chr14_122104619_122105909 | 1.05 |
A330035P11Rik |
RIKEN cDNA A330035P11 gene |
1689 |
0.23 |
| chr9_7763414_7764605 | 1.02 |
Tmem123 |
transmembrane protein 123 |
32 |
0.97 |
| chrX_53643664_53643984 | 1.02 |
Rtl8a |
retrotransposon Gag like 8A |
142 |
0.92 |
| chr7_27310306_27310823 | 1.01 |
Ltbp4 |
latent transforming growth factor beta binding protein 4 |
95 |
0.94 |
| chr7_15954917_15955481 | 1.01 |
Nop53 |
NOP53 ribosome biogenesis factor |
9125 |
0.1 |
| chrX_136957917_136958236 | 1.00 |
Tmsb15b2 |
thymosin beta 15b2 |
51 |
0.94 |
| chr9_21797681_21798988 | 1.00 |
Kank2 |
KN motif and ankyrin repeat domains 2 |
169 |
0.91 |
| chr17_56424982_56425554 | 1.00 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
135 |
0.95 |
| chr16_90220138_90221417 | 0.97 |
Sod1 |
superoxide dismutase 1, soluble |
23 |
0.97 |
| chr4_133055540_133056727 | 0.97 |
Ahdc1 |
AT hook, DNA binding motif, containing 1 |
10116 |
0.19 |
| chr6_88841841_88842983 | 0.97 |
Abtb1 |
ankyrin repeat and BTB (POZ) domain containing 1 |
428 |
0.42 |
| chr4_63216287_63217000 | 0.97 |
Col27a1 |
collagen, type XXVII, alpha 1 |
1208 |
0.45 |
| chr10_87057937_87058923 | 0.96 |
1700113H08Rik |
RIKEN cDNA 1700113H08 gene |
384 |
0.87 |
| chr9_107714405_107714990 | 0.96 |
Sema3f |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
4222 |
0.11 |
| chr6_88942621_88943289 | 0.96 |
4933427D06Rik |
RIKEN cDNA 4933427D06 gene |
7728 |
0.14 |
| chr13_107372004_107372155 | 0.95 |
Gm2736 |
predicted gene 2736 |
23875 |
0.19 |
| chr10_81383963_81384933 | 0.94 |
Dohh |
deoxyhypusine hydroxylase/monooxygenase |
11 |
0.48 |
| chr15_99771905_99772913 | 0.94 |
Cers5 |
ceramide synthase 5 |
53 |
0.94 |
| chr2_33130296_33131698 | 0.93 |
Garnl3 |
GTPase activating RANGAP domain-like 3 |
389 |
0.84 |
| chr7_27256966_27257266 | 0.93 |
Gm15567 |
predicted gene 15567 |
816 |
0.33 |
| chr5_142551306_142551542 | 0.92 |
Radil |
Ras association and DIL domains |
326 |
0.85 |
| chr4_45018456_45019009 | 0.92 |
Polr1e |
polymerase (RNA) I polypeptide E |
35 |
0.96 |
| chr17_51790725_51791001 | 0.91 |
5830444F18Rik |
RIKEN cDNA 5830444F18 gene |
2598 |
0.16 |
| chr13_98354125_98354312 | 0.90 |
Foxd1 |
forkhead box D1 |
24 |
0.97 |
| chr7_100932502_100932992 | 0.89 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
640 |
0.64 |
| chr8_83667823_83669013 | 0.89 |
Ptger1 |
prostaglandin E receptor 1 (subtype EP1) |
1276 |
0.29 |
| chr12_102554281_102554622 | 0.89 |
Chga |
chromogranin A |
518 |
0.75 |
| chr9_107526565_107527044 | 0.89 |
Cacna2d2 |
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
147 |
0.87 |
| chr2_158794072_158795253 | 0.88 |
Dhx35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
145 |
0.96 |
| chr5_4623720_4623923 | 0.88 |
Fzd1 |
frizzled class receptor 1 |
134214 |
0.04 |
| chr15_76538707_76539267 | 0.87 |
Slc52a2 |
solute carrier protein 52, member 2 |
23 |
0.74 |
| chr18_46597470_46598409 | 0.87 |
Eif1a |
eukaryotic translation initiation factor 1A |
75 |
0.84 |
| chr5_37241363_37242150 | 0.86 |
Crmp1 |
collapsin response mediator protein 1 |
184 |
0.95 |
| chr2_160730613_160731328 | 0.86 |
Plcg1 |
phospholipase C, gamma 1 |
330 |
0.87 |
| chr7_19010518_19010930 | 0.86 |
Irf2bp1 |
interferon regulatory factor 2 binding protein 1 |
6680 |
0.07 |
| chr5_35757643_35758570 | 0.84 |
Ablim2 |
actin-binding LIM protein 2 |
55 |
0.97 |
| chr8_3515797_3516665 | 0.84 |
Pnpla6 |
patatin-like phospholipase domain containing 6 |
430 |
0.7 |
| chr9_121403105_121403296 | 0.84 |
Trak1 |
trafficking protein, kinesin binding 1 |
278 |
0.91 |
| chr9_22050523_22050944 | 0.84 |
Elavl3 |
ELAV like RNA binding protein 3 |
1277 |
0.23 |
| chr12_108554100_108554849 | 0.84 |
Evl |
Ena-vasodilator stimulated phosphoprotein |
246 |
0.85 |
| chr1_21161877_21162028 | 0.83 |
Gm2693 |
predicted gene 2693 |
17036 |
0.15 |
| chr10_81167633_81168074 | 0.82 |
Pias4 |
protein inhibitor of activated STAT 4 |
70 |
0.92 |
| chr17_56757443_56758083 | 0.82 |
Nrtn |
neurturin |
233 |
0.86 |
| chr3_20366868_20367395 | 0.81 |
Agtr1b |
angiotensin II receptor, type 1b |
7 |
0.98 |
| chr16_42339557_42339849 | 0.81 |
Gap43 |
growth associated protein 43 |
948 |
0.66 |
| chr15_76987616_76987801 | 0.80 |
Gm35933 |
predicted gene, 35933 |
316 |
0.82 |
| chr8_70315603_70316677 | 0.80 |
Cers1 |
ceramide synthase 1 |
353 |
0.75 |
| chr13_28946898_28947096 | 0.80 |
Sox4 |
SRY (sex determining region Y)-box 4 |
6716 |
0.22 |
| chr16_13818843_13819686 | 0.80 |
Ntan1 |
N-terminal Asn amidase |
13 |
0.5 |
| chr18_49979102_49979704 | 0.80 |
Tnfaip8 |
tumor necrosis factor, alpha-induced protein 8 |
24 |
0.98 |
| chr4_9843986_9844421 | 0.79 |
Gdf6 |
growth differentiation factor 6 |
169 |
0.96 |
| chr3_104794518_104794903 | 0.79 |
Rhoc |
ras homolog family member C |
2727 |
0.13 |
| chr10_81464661_81465630 | 0.79 |
Gm16105 |
predicted gene 16105 |
3986 |
0.08 |
| chr7_107594304_107595463 | 0.78 |
Ppfibp2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
324 |
0.83 |
| chr4_129440796_129440989 | 0.78 |
Zbtb8b |
zinc finger and BTB domain containing 8b |
39 |
0.96 |
| chr13_92530160_92531475 | 0.77 |
Zfyve16 |
zinc finger, FYVE domain containing 16 |
51 |
0.98 |
| chr10_42477881_42479089 | 0.77 |
Afg1l |
AFG1 like ATPase |
70 |
0.97 |
| chr1_125560780_125561505 | 0.77 |
Slc35f5 |
solute carrier family 35, member F5 |
126 |
0.98 |
| chr8_69822383_69823195 | 0.76 |
Lpar2 |
lysophosphatidic acid receptor 2 |
205 |
0.88 |
| chr13_98009130_98009281 | 0.75 |
Arhgef28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
7212 |
0.28 |
| chr17_56078902_56079299 | 0.75 |
Hdgfl2 |
HDGF like 2 |
534 |
0.58 |
| chr4_32656940_32657186 | 0.74 |
Mdn1 |
midasin AAA ATPase 1 |
56 |
0.94 |
| chr3_90509125_90509750 | 0.74 |
Chtop |
chromatin target of PRMT1 |
13 |
0.93 |
| chr15_101221336_101221785 | 0.74 |
AU021063 |
expressed sequence AU021063 |
313 |
0.78 |
| chr5_138279873_138280191 | 0.74 |
Gpc2 |
glypican 2 (cerebroglycan) |
27 |
0.44 |
| chr16_18686655_18686806 | 0.74 |
Rps2-ps7 |
ribosomal protein S2, pseudogene 7 |
4514 |
0.18 |
| chr5_32785233_32785830 | 0.74 |
Pisd |
phosphatidylserine decarboxylase |
50 |
0.96 |
| chr7_132896569_132896720 | 0.73 |
1500002F19Rik |
RIKEN cDNA 1500002F19 gene |
7957 |
0.14 |
| chr15_11995897_11996209 | 0.71 |
Sub1 |
SUB1 homolog, transcriptional regulator |
5 |
0.98 |
| chr2_148732332_148732639 | 0.71 |
Napb |
N-ethylmaleimide sensitive fusion protein attachment protein beta |
18 |
0.97 |
| chr15_31338807_31338958 | 0.71 |
Ankrd33b |
ankyrin repeat domain 33B |
13633 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.9 | 2.8 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.5 | 2.6 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.5 | 2.0 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.5 | 1.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.4 | 1.3 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.4 | 1.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 0.7 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.3 | 1.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.3 | 0.7 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 2.8 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 0.9 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.3 | 3.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 | 1.2 | GO:1903977 | positive regulation of glial cell migration(GO:1903977) |
| 0.3 | 0.9 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.3 | 1.8 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.3 | 1.3 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 2.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 1.8 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.2 | 0.9 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 0.7 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 0.7 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 1.1 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.2 | 3.1 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 0.6 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 | 1.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 0.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 0.6 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.2 | 9.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 0.6 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.2 | 2.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.2 | 0.5 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.2 | 0.9 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.2 | 0.5 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 0.6 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.2 | 0.8 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 1.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.4 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 1.9 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.1 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.4 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.4 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 2.8 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.5 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 1.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.4 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 1.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 1.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.3 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 | 0.2 | GO:0045632 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.1 | 0.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 3.9 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.2 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.4 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 1.1 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.6 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.8 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.1 | 0.3 | GO:0060066 | oviduct development(GO:0060066) |
| 0.1 | 0.2 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) |
| 0.1 | 0.3 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.3 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.1 | 0.3 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.1 | 0.3 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 1.4 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 1.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.7 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.2 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.1 | 0.2 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.1 | 0.2 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.1 | 0.3 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 0.4 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.1 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.1 | 0.4 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 0.4 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.1 | 1.0 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.2 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.6 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.2 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 | 0.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 1.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.1 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 2.0 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.4 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.2 | GO:1904751 | regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.0 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 0.5 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 1.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.2 | GO:0042504 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) |
| 0.1 | 0.5 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.1 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.1 | GO:0021636 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.1 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.2 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.1 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.0 | 0.4 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0002254 | kinin cascade(GO:0002254) |
| 0.0 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.3 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.5 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.5 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.5 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.3 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.2 | GO:0051198 | negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.0 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.2 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.0 | 0.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.2 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:1901524 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.7 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.3 | GO:2000018 | regulation of male gonad development(GO:2000018) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.5 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 2.0 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.1 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:1902267 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 | 0.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 1.8 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.7 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.1 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.3 | GO:0033866 | nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 0.4 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.0 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 | 0.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.2 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0010878 | cholesterol storage(GO:0010878) |
| 0.0 | 0.1 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.5 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.9 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 1.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.0 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.3 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.6 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.3 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.0 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.0 | GO:0060405 | regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.0 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.0 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 0.0 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.7 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 | 0.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.0 | GO:0052200 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.0 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.0 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.6 | GO:0015844 | monoamine transport(GO:0015844) |
| 0.0 | 0.1 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.0 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.0 | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.0 | 0.0 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.0 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.0 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 | 0.0 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.5 | 1.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.4 | 1.3 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.4 | 2.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 2.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.3 | 1.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 0.7 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 1.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 0.4 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.2 | 0.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.2 | 1.9 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 0.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 0.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 0.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 1.9 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 0.8 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.8 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.8 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 1.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.6 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 1.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 4.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 5.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.8 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.4 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 3.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.2 | GO:0033647 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.1 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 2.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.7 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0000802 | transverse filament(GO:0000802) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.0 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.0 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.4 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.0 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.2 | GO:0044298 | cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 5.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:0009279 | cell outer membrane(GO:0009279) cell envelope(GO:0030313) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 1.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 10.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.7 | 2.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.7 | 3.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.6 | 1.8 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) neuroligin family protein binding(GO:0097109) |
| 0.6 | 2.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.4 | 1.2 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.4 | 1.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.4 | 1.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 2.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.3 | 0.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 1.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 0.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 1.8 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 3.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 0.9 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 1.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.5 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 0.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 0.8 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 2.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.4 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.1 | 1.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 1.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.4 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.1 | 1.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.3 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 2.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 1.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.4 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.1 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.2 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.1 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.4 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.3 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.5 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.6 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.6 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 1.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 2.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0016019 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0043829 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.4 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 5.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.0 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0031559 | oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.2 | GO:0034882 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 3.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.4 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.4 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 0.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 2.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 2.7 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 2.0 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 1.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 2.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.7 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.1 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 2.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |