Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
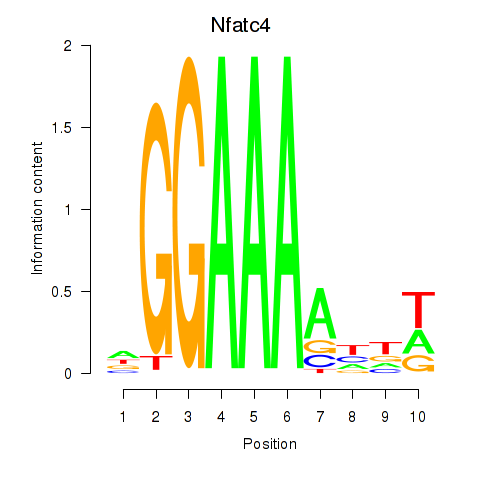
Results for Nfatc4
Z-value: 0.35
Transcription factors associated with Nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc4
|
ENSMUSG00000023411.5 | nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 4 |
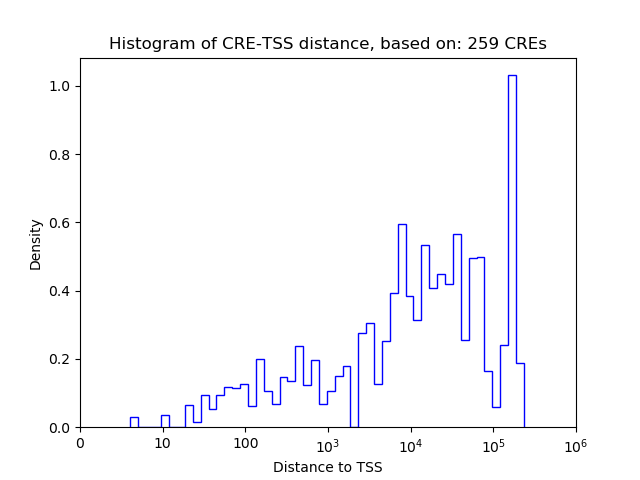
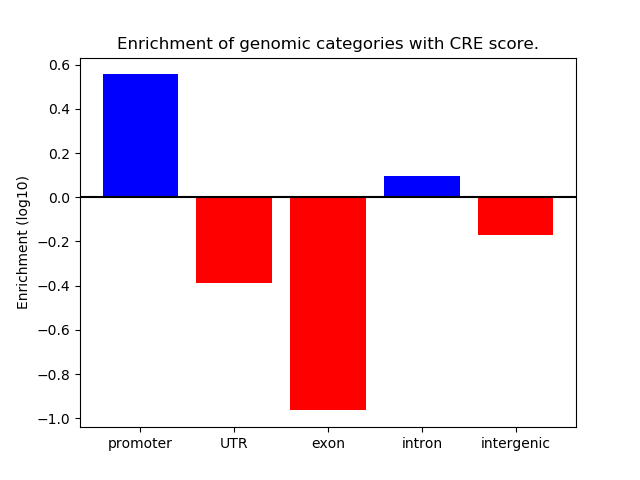
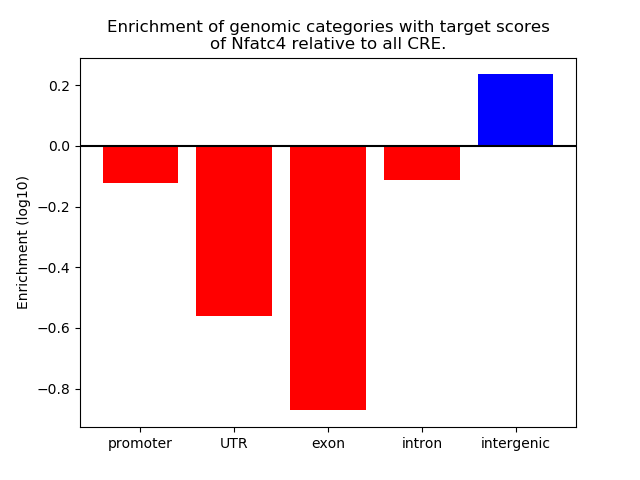
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_55824498_55825973 | Nfatc4 | 198 | 0.874152 | 0.33 | 1.5e-02 | Click! |
| chr14_55824260_55824444 | Nfatc4 | 443 | 0.669096 | 0.32 | 1.7e-02 | Click! |
| chr14_55825986_55826137 | Nfatc4 | 260 | 0.827506 | 0.29 | 3.0e-02 | Click! |
| chr14_55822948_55823744 | Nfatc4 | 202 | 0.870721 | 0.29 | 3.3e-02 | Click! |
| chr14_55823772_55824002 | Nfatc4 | 743 | 0.452656 | 0.28 | 4.2e-02 | Click! |
Activity of the Nfatc4 motif across conditions
Conditions sorted by the z-value of the Nfatc4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
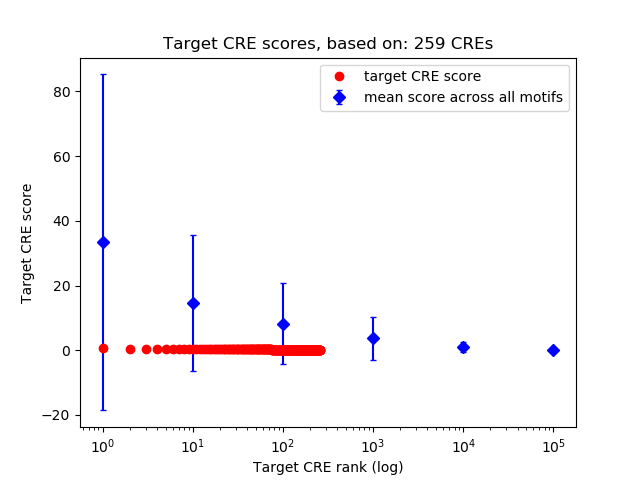
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_68595404_68595555 | 0.56 |
Brinp1 |
bone morphogenic protein/retinoic acid inducible neural specific 1 |
233879 |
0.02 |
| chr4_23173209_23173360 | 0.53 |
Gm11884 |
predicted gene 11884 |
87500 |
0.1 |
| chr10_101317018_101317229 | 0.52 |
Gm19233 |
predicted gene, 19233 |
45502 |
0.2 |
| chr3_117002231_117002410 | 0.50 |
Gm9332 |
predicted gene 9332 |
2734 |
0.23 |
| chr13_111237965_111238337 | 0.50 |
3110015C05Rik |
RIKEN cDNA 3110015C05 gene |
8720 |
0.21 |
| chr8_55121794_55122373 | 0.48 |
Gm8734 |
predicted gene 8734 |
40368 |
0.14 |
| chr2_109694445_109694667 | 0.48 |
Bdnf |
brain derived neurotrophic factor |
87 |
0.97 |
| chr1_186505235_186505386 | 0.48 |
A730004F24Rik |
RIKEN cDNA A730004F24 gene |
53162 |
0.15 |
| chr8_55371986_55372445 | 0.48 |
Gm20586 |
predicted gene, 20586 |
160076 |
0.04 |
| chr3_70594904_70595055 | 0.46 |
Gm37213 |
predicted gene, 37213 |
27683 |
0.24 |
| chr8_55358672_55359287 | 0.45 |
Gm20586 |
predicted gene, 20586 |
173312 |
0.03 |
| chr8_55352073_55352667 | 0.44 |
Gm20586 |
predicted gene, 20586 |
179921 |
0.03 |
| chr2_22721578_22721729 | 0.42 |
Gm13338 |
predicted gene 13338 |
8142 |
0.15 |
| chr1_24226131_24226533 | 0.42 |
Col9a1 |
collagen, type IX, alpha 1 |
3185 |
0.33 |
| chr8_55355373_55355937 | 0.42 |
Gm20586 |
predicted gene, 20586 |
176636 |
0.03 |
| chr2_97467028_97467474 | 0.41 |
Lrrc4c |
leucine rich repeat containing 4C |
406 |
0.92 |
| chr8_55115169_55115732 | 0.40 |
Gm8734 |
predicted gene 8734 |
33735 |
0.14 |
| chr15_53370344_53370653 | 0.40 |
Gm27926 |
predicted gene, 27926 |
13421 |
0.24 |
| chr2_12970544_12970695 | 0.40 |
Pter |
phosphotriesterase related |
7320 |
0.17 |
| chr3_102366187_102366567 | 0.40 |
Gm43242 |
predicted gene 43242 |
2785 |
0.27 |
| chr2_147702775_147702949 | 0.39 |
A530006G24Rik |
RIKEN cDNA A530006G24 gene |
7831 |
0.23 |
| chr2_66409856_66410177 | 0.39 |
Scn1a |
sodium channel, voltage-gated, type I, alpha |
48 |
0.9 |
| chr1_98047379_98047565 | 0.39 |
B230216N24Rik |
RIKEN cDNA B230216N24 gene |
282 |
0.89 |
| chr6_30552193_30552508 | 0.39 |
Cpa2 |
carboxypeptidase A2, pancreatic |
447 |
0.74 |
| chr9_91350886_91351318 | 0.38 |
A730094K22Rik |
RIKEN cDNA A730094K22 gene |
149 |
0.92 |
| chr16_88681405_88681556 | 0.38 |
Gm7667 |
predicted gene 7667 |
1248 |
0.3 |
| chr14_7749794_7749966 | 0.38 |
Gm16741 |
predicted gene, 16741 |
65 |
0.97 |
| chr4_119460677_119461253 | 0.37 |
Zmynd12 |
zinc finger, MYND domain containing 12 |
16245 |
0.1 |
| chr8_55349027_55349338 | 0.37 |
Gm20586 |
predicted gene, 20586 |
183109 |
0.03 |
| chr3_34443838_34444057 | 0.37 |
Gm20515 |
predicted gene 20515 |
8850 |
0.18 |
| chr15_6934781_6934981 | 0.36 |
Osmr |
oncostatin M receptor |
59912 |
0.13 |
| chr3_142411042_142411193 | 0.34 |
Pdlim5 |
PDZ and LIM domain 5 |
15421 |
0.23 |
| chr2_63963866_63964056 | 0.34 |
Fign |
fidgetin |
134027 |
0.06 |
| chr4_27988130_27988413 | 0.33 |
Tpm3-rs2 |
tropomyosin 3, related sequence 2 |
8634 |
0.29 |
| chr2_58135097_58135248 | 0.32 |
Cytip |
cytohesin 1 interacting protein |
21625 |
0.16 |
| chr14_70893868_70894196 | 0.32 |
Gfra2 |
glial cell line derived neurotrophic factor family receptor alpha 2 |
3504 |
0.28 |
| chr8_55362331_55362582 | 0.32 |
Gm20586 |
predicted gene, 20586 |
169835 |
0.03 |
| chr14_122324157_122324364 | 0.31 |
Gm25464 |
predicted gene, 25464 |
63900 |
0.09 |
| chr15_54312323_54312474 | 0.31 |
Gm5215 |
predicted gene 5215 |
33 |
0.98 |
| chr10_4553737_4553991 | 0.31 |
Gm3266 |
predicted gene 3266 |
16359 |
0.17 |
| chr13_25754626_25754869 | 0.30 |
Gm11350 |
predicted gene 11350 |
73604 |
0.12 |
| chr3_145645937_145646121 | 0.30 |
Ccn1 |
cellular communication network factor 1 |
3952 |
0.21 |
| chr13_78795499_78795660 | 0.30 |
Gm48402 |
predicted gene, 48402 |
157562 |
0.04 |
| chr15_27768039_27768190 | 0.30 |
Trio |
triple functional domain (PTPRF interacting) |
19563 |
0.21 |
| chr2_116894201_116894398 | 0.29 |
D330050G23Rik |
RIKEN cDNA D330050G23 gene |
5853 |
0.19 |
| chr3_18905759_18906119 | 0.29 |
Gm30341 |
predicted gene, 30341 |
152862 |
0.04 |
| chr10_56378893_56379326 | 0.28 |
Gja1 |
gap junction protein, alpha 1 |
21 |
0.98 |
| chr12_30129274_30129604 | 0.28 |
Tpo |
thyroid peroxidase |
2460 |
0.37 |
| chr8_55118866_55119053 | 0.28 |
Gm8734 |
predicted gene 8734 |
37244 |
0.14 |
| chrX_110077953_110078105 | 0.28 |
Gm4991 |
predicted gene 4991 |
167808 |
0.04 |
| chr5_104416083_104416237 | 0.27 |
Spp1 |
secreted phosphoprotein 1 |
18958 |
0.15 |
| chr15_57059947_57060098 | 0.27 |
Gm2582 |
predicted gene 2582 |
53398 |
0.17 |
| chr11_102603589_102603785 | 0.27 |
Fzd2 |
frizzled class receptor 2 |
709 |
0.5 |
| chr8_55368995_55369146 | 0.27 |
Gm20586 |
predicted gene, 20586 |
163221 |
0.04 |
| chr8_55345851_55346060 | 0.27 |
Gm20586 |
predicted gene, 20586 |
186336 |
0.03 |
| chr4_85486606_85486757 | 0.27 |
Adamtsl1 |
ADAMTS-like 1 |
27491 |
0.25 |
| chr2_13438384_13438535 | 0.26 |
Cubn |
cubilin (intrinsic factor-cobalamin receptor) |
17168 |
0.25 |
| chr13_91187692_91187893 | 0.26 |
Gm17450 |
predicted gene, 17450 |
32660 |
0.16 |
| chr1_168908656_168908844 | 0.25 |
Mir6354 |
microRNA 6354 |
110339 |
0.07 |
| chr14_123065129_123065306 | 0.25 |
AA536875 |
expressed sequence AA536875 |
21935 |
0.22 |
| chr1_30701244_30701419 | 0.25 |
Gm9898 |
predicted gene 9898 |
18631 |
0.22 |
| chrX_150955690_150955875 | 0.25 |
Gm8424 |
predicted gene 8424 |
29623 |
0.14 |
| chr15_26895841_26896059 | 0.25 |
Fbxl7 |
F-box and leucine-rich repeat protein 7 |
370 |
0.92 |
| chr15_48790820_48790999 | 0.24 |
Csmd3 |
CUB and Sushi multiple domains 3 |
1024 |
0.66 |
| chr3_13472076_13472230 | 0.24 |
Gm2464 |
predicted gene 2464 |
324 |
0.62 |
| chr15_83848303_83848454 | 0.24 |
Mpped1 |
metallophosphoesterase domain containing 1 |
12338 |
0.22 |
| chr6_137699179_137699330 | 0.24 |
Strap |
serine/threonine kinase receptor associated protein |
35824 |
0.16 |
| chr12_116485218_116485891 | 0.23 |
Ptprn2 |
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
166 |
0.93 |
| chr10_37139778_37139969 | 0.23 |
5930403N24Rik |
RIKEN cDNA 5930403N24 gene |
41 |
0.95 |
| chr18_21728656_21729116 | 0.23 |
Klhl14 |
kelch-like 14 |
74168 |
0.11 |
| chr2_102094776_102094927 | 0.23 |
Gm13920 |
predicted gene 13920 |
37264 |
0.14 |
| chr5_49350667_49350890 | 0.23 |
Gm42772 |
predicted gene 42772 |
61054 |
0.09 |
| chr12_100763079_100763279 | 0.22 |
Gm25801 |
predicted gene, 25801 |
5949 |
0.16 |
| chr4_90438582_90438776 | 0.22 |
Gm12635 |
predicted gene 12635 |
14195 |
0.24 |
| chr1_158163507_158163658 | 0.22 |
Brinp2 |
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
132882 |
0.05 |
| chr9_82044648_82044799 | 0.22 |
Gm37837 |
predicted gene, 37837 |
16143 |
0.26 |
| chr8_90536167_90536332 | 0.22 |
Gm45639 |
predicted gene 45639 |
138709 |
0.04 |
| chr11_76912041_76912192 | 0.22 |
Tmigd1 |
transmembrane and immunoglobulin domain containing 1 |
7554 |
0.17 |
| chr1_85870211_85870362 | 0.21 |
Itm2c |
integral membrane protein 2C |
23995 |
0.12 |
| chr6_125163930_125164575 | 0.21 |
Gapdh |
glyceraldehyde-3-phosphate dehydrogenase |
664 |
0.41 |
| chr13_89669404_89669574 | 0.21 |
Gm16318 |
predicted gene 16318 |
6719 |
0.21 |
| chr13_31792557_31792841 | 0.21 |
Gm11379 |
predicted gene 11379 |
4840 |
0.19 |
| chr8_55378818_55378969 | 0.21 |
Gm20586 |
predicted gene, 20586 |
153398 |
0.04 |
| chr10_29217677_29217828 | 0.21 |
9330159F19Rik |
RIKEN cDNA 9330159F19 gene |
6047 |
0.18 |
| chr8_88266957_88267125 | 0.21 |
Adcy7 |
adenylate cyclase 7 |
5362 |
0.2 |
| chr15_21407429_21407584 | 0.20 |
Gm38234 |
predicted gene, 38234 |
227349 |
0.02 |
| chr5_125149190_125149438 | 0.20 |
Ncor2 |
nuclear receptor co-repressor 2 |
24414 |
0.18 |
| chr4_13335515_13335743 | 0.20 |
Gm26250 |
predicted gene, 26250 |
68579 |
0.12 |
| chr2_110565079_110565282 | 0.20 |
Gm10794 |
predicted gene 10794 |
31457 |
0.18 |
| chr18_7535204_7535355 | 0.20 |
Gm18948 |
predicted gene, 18948 |
13708 |
0.23 |
| chr11_29448855_29449034 | 0.20 |
Ccdc88a |
coiled coil domain containing 88A |
7552 |
0.17 |
| chr5_15043429_15043756 | 0.20 |
Gm17019 |
predicted gene 17019 |
10594 |
0.25 |
| chr17_44398035_44398193 | 0.20 |
Gm49872 |
predicted gene, 49872 |
30211 |
0.23 |
| chr8_7131824_7132038 | 0.20 |
Gm44910 |
predicted gene 44910 |
1033 |
0.65 |
| chr11_72042565_72042858 | 0.19 |
Pimreg |
PICALM interacting mitotic regulator |
187 |
0.93 |
| chr15_67944519_67944685 | 0.19 |
Gm49409 |
predicted gene, 49409 |
1798 |
0.49 |
| chr5_141139181_141139332 | 0.19 |
Gm26113 |
predicted gene, 26113 |
58825 |
0.12 |
| chr4_84018889_84019101 | 0.19 |
6030471H07Rik |
RIKEN cDNA 6030471H07 gene |
25884 |
0.22 |
| chr17_21732990_21733141 | 0.19 |
Zfp229 |
zinc finger protein 229 |
615 |
0.62 |
| chr16_77422519_77422863 | 0.19 |
9430053O09Rik |
RIKEN cDNA 9430053O09 gene |
871 |
0.45 |
| chr15_52161086_52161419 | 0.19 |
Gm2387 |
predicted gene 2387 |
71944 |
0.1 |
| chr1_26282913_26283064 | 0.19 |
Gm19123 |
predicted gene, 19123 |
183248 |
0.03 |
| chr2_166517600_166517812 | 0.19 |
Gm14267 |
predicted gene 14267 |
1340 |
0.5 |
| chr10_13324534_13324685 | 0.19 |
Phactr2 |
phosphatase and actin regulator 2 |
320 |
0.93 |
| chr2_104219978_104220129 | 0.18 |
Gm22501 |
predicted gene, 22501 |
8185 |
0.16 |
| chr6_4739869_4740257 | 0.18 |
Sgce |
sarcoglycan, epsilon |
144 |
0.95 |
| chr18_89564398_89564718 | 0.18 |
Dok6 |
docking protein 6 |
4489 |
0.24 |
| chr5_131645868_131646247 | 0.18 |
Gm43483 |
predicted gene 43483 |
3250 |
0.16 |
| chr3_115619961_115620260 | 0.18 |
S1pr1 |
sphingosine-1-phosphate receptor 1 |
94962 |
0.07 |
| chr16_14445264_14445415 | 0.18 |
Abcc1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
15669 |
0.17 |
| chr4_49232759_49232939 | 0.18 |
Gm22685 |
predicted gene, 22685 |
56449 |
0.13 |
| chr9_122836785_122837126 | 0.18 |
Gm35549 |
predicted gene, 35549 |
6931 |
0.11 |
| chr3_27862930_27863950 | 0.17 |
Gm26040 |
predicted gene, 26040 |
4673 |
0.26 |
| chr1_156782534_156782685 | 0.17 |
Gm15428 |
predicted pseudogene 15428 |
10903 |
0.15 |
| chr2_97543385_97543550 | 0.17 |
Lrrc4c |
leucine rich repeat containing 4C |
75378 |
0.11 |
| chr12_112466043_112466390 | 0.17 |
A530016L24Rik |
RIKEN cDNA A530016L24 gene |
23232 |
0.13 |
| chrX_43430637_43430950 | 0.17 |
Tenm1 |
teneurin transmembrane protein 1 |
1667 |
0.41 |
| chr10_27132998_27133149 | 0.17 |
Lama2 |
laminin, alpha 2 |
76428 |
0.1 |
| chr1_78745515_78745771 | 0.17 |
Gm6159 |
predicted gene 6159 |
39216 |
0.13 |
| chr6_40226222_40226373 | 0.17 |
Gm37003 |
predicted gene, 37003 |
78 |
0.97 |
| chr13_113495968_113496190 | 0.17 |
4921509O07Rik |
RIKEN cDNA 4921509O07 gene |
6765 |
0.16 |
| chr5_14949744_14950125 | 0.17 |
Speer4e |
spermatogenesis associated glutamate (E)-rich protein 4e |
11505 |
0.19 |
| chr15_89148579_89149159 | 0.16 |
Mapk11 |
mitogen-activated protein kinase 11 |
625 |
0.51 |
| chr9_39604057_39604411 | 0.16 |
AW551984 |
expressed sequence AW551984 |
110 |
0.93 |
| chr6_127011987_127012359 | 0.16 |
Fgf6 |
fibroblast growth factor 6 |
3413 |
0.16 |
| chr10_26711151_26711302 | 0.16 |
Gm48893 |
predicted gene, 48893 |
15652 |
0.15 |
| chr6_108235594_108236116 | 0.16 |
Itpr1 |
inositol 1,4,5-trisphosphate receptor 1 |
17103 |
0.23 |
| chr16_34228583_34229181 | 0.15 |
Kalrn |
kalirin, RhoGEF kinase |
23433 |
0.27 |
| chr16_80512891_80513042 | 0.15 |
Gm24773 |
predicted gene, 24773 |
172259 |
0.03 |
| chr6_142702177_142702435 | 0.15 |
Abcc9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
9 |
0.98 |
| chr10_42477881_42479089 | 0.15 |
Afg1l |
AFG1 like ATPase |
70 |
0.97 |
| chr7_6753498_6753708 | 0.15 |
Gm44863 |
predicted gene 44863 |
560 |
0.61 |
| chr14_75206107_75206258 | 0.15 |
Gm15629 |
predicted gene 15629 |
6911 |
0.15 |
| chr19_52048467_52048668 | 0.15 |
Gm50330 |
predicted gene, 50330 |
128234 |
0.05 |
| chr16_56920328_56920584 | 0.15 |
Lnp1 |
leukemia NUP98 fusion partner 1 |
7605 |
0.16 |
| chr3_145560223_145560406 | 0.15 |
Col24a1 |
collagen, type XXIV, alpha 1 |
15341 |
0.19 |
| chr13_47223822_47224099 | 0.14 |
Rnf144b |
ring finger protein 144B |
30135 |
0.16 |
| chr11_85676422_85676576 | 0.14 |
Bcas3 |
breast carcinoma amplified sequence 3 |
35795 |
0.15 |
| chr1_78816664_78817000 | 0.14 |
Kcne4 |
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
74 |
0.97 |
| chr13_48228069_48228495 | 0.14 |
Gm36346 |
predicted gene, 36346 |
8969 |
0.16 |
| chr3_109025491_109025897 | 0.14 |
Fam102b |
family with sequence similarity 102, member B |
1579 |
0.31 |
| chr11_110251644_110251914 | 0.13 |
Abca6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
3 |
0.99 |
| chr13_59850542_59850693 | 0.13 |
Gm48384 |
predicted gene, 48384 |
23025 |
0.1 |
| chr19_26745483_26745907 | 0.13 |
Smarca2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
486 |
0.85 |
| chr4_40837444_40837899 | 0.13 |
Mir5123 |
microRNA 5123 |
12467 |
0.11 |
| chr2_125355381_125355540 | 0.13 |
Gm14003 |
predicted gene 14003 |
71482 |
0.09 |
| chr3_157021687_157021992 | 0.13 |
Gm43527 |
predicted gene 43527 |
21422 |
0.21 |
| chr4_43561783_43562397 | 0.12 |
Tln1 |
talin 1 |
67 |
0.71 |
| chr4_53011544_53011738 | 0.12 |
Nipsnap3b |
nipsnap homolog 3B |
239 |
0.91 |
| chr17_49703998_49704149 | 0.12 |
Kif6 |
kinesin family member 6 |
10037 |
0.27 |
| chr6_33929843_33930025 | 0.12 |
Gm13853 |
predicted gene 13853 |
62477 |
0.12 |
| chr1_162144363_162144537 | 0.12 |
Dnm3 |
dynamin 3 |
9698 |
0.2 |
| chr16_31575682_31575833 | 0.12 |
Gm34256 |
predicted gene, 34256 |
17463 |
0.16 |
| chr11_98412973_98413218 | 0.12 |
Erbb2 |
erb-b2 receptor tyrosine kinase 2 |
602 |
0.51 |
| chr5_97953119_97953468 | 0.12 |
Antxr2 |
anthrax toxin receptor 2 |
42802 |
0.14 |
| chr3_69285315_69285466 | 0.12 |
Ppm1l |
protein phosphatase 1 (formerly 2C)-like |
31471 |
0.14 |
| chr3_8509688_8509990 | 0.12 |
Stmn2 |
stathmin-like 2 |
253 |
0.93 |
| chr1_45898190_45898341 | 0.12 |
Gm18303 |
predicted gene, 18303 |
7358 |
0.14 |
| chr5_85795902_85796137 | 0.11 |
Gm24626 |
predicted gene, 24626 |
33608 |
0.21 |
| chr15_26880673_26880848 | 0.11 |
Fbxl7 |
F-box and leucine-rich repeat protein 7 |
14758 |
0.26 |
| chr2_63963398_63963819 | 0.11 |
Fign |
fidgetin |
134380 |
0.06 |
| chr1_39902764_39903132 | 0.11 |
Map4k4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
1718 |
0.42 |
| chr9_55429377_55429605 | 0.11 |
Etfa |
electron transferring flavoprotein, alpha polypeptide |
58022 |
0.1 |
| chr1_193086803_193086954 | 0.11 |
Utp25 |
UTP25 small subunit processome component |
27797 |
0.12 |
| chr8_110716047_110716632 | 0.11 |
Mtss2 |
MTSS I-BAR domain containing 2 |
5137 |
0.18 |
| chrX_130452263_130452432 | 0.11 |
Gm14984 |
predicted gene 14984 |
50128 |
0.14 |
| chr4_49213275_49213426 | 0.11 |
Gm22685 |
predicted gene, 22685 |
75948 |
0.09 |
| chr1_30719021_30719172 | 0.10 |
Gm9898 |
predicted gene 9898 |
866 |
0.65 |
| chr3_148992115_148992398 | 0.10 |
Adgrl2 |
adhesion G protein-coupled receptor L2 |
1701 |
0.32 |
| chr7_63924745_63925223 | 0.10 |
Klf13 |
Kruppel-like factor 13 |
114 |
0.95 |
| chrX_109845362_109845527 | 0.10 |
Gm4991 |
predicted gene 4991 |
64777 |
0.15 |
| chr1_67226374_67226525 | 0.10 |
Gm15668 |
predicted gene 15668 |
22751 |
0.2 |
| chr2_177498924_177499075 | 0.10 |
Gm14403 |
predicted gene 14403 |
684 |
0.66 |
| chr7_116736401_116736714 | 0.10 |
Gm25423 |
predicted gene, 25423 |
45072 |
0.15 |
| chr8_10318195_10318346 | 0.09 |
Myo16 |
myosin XVI |
45698 |
0.18 |
| chrX_133911740_133912091 | 0.09 |
Srpx2 |
sushi-repeat-containing protein, X-linked 2 |
3467 |
0.2 |
| chr5_34252384_34252580 | 0.09 |
Zfyve28 |
zinc finger, FYVE domain containing 28 |
8923 |
0.12 |
| chr2_6928083_6928276 | 0.09 |
Gm10855 |
predicted gene 10855 |
6902 |
0.18 |
| chr6_127150352_127150816 | 0.09 |
Ccnd2 |
cyclin D2 |
377 |
0.68 |
| chr7_112289842_112290024 | 0.09 |
Mical2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
18634 |
0.26 |
| chr2_85123274_85123495 | 0.09 |
Aplnr |
apelin receptor |
12841 |
0.12 |
| chr2_72886288_72886551 | 0.08 |
Sp3 |
trans-acting transcription factor 3 |
54665 |
0.1 |
| chr6_100707892_100708043 | 0.08 |
Gxylt2 |
glucoside xylosyltransferase 2 |
3212 |
0.21 |
| chr12_31654748_31655394 | 0.08 |
Cog5 |
component of oligomeric golgi complex 5 |
202 |
0.54 |
| chr1_134858695_134859022 | 0.08 |
Ppp1r12b |
protein phosphatase 1, regulatory subunit 12B |
12438 |
0.15 |
| chr4_149606130_149606281 | 0.08 |
Clstn1 |
calsyntenin 1 |
18957 |
0.12 |
| chr18_62804016_62804176 | 0.08 |
2700046A07Rik |
RIKEN cDNA 2700046A07 gene |
47749 |
0.14 |
| chr7_100925122_100925295 | 0.08 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
6899 |
0.15 |
| chr13_70803034_70803185 | 0.08 |
Gm36529 |
predicted gene, 36529 |
37342 |
0.15 |
| chr1_57850016_57850167 | 0.08 |
Spats2l |
spermatogenesis associated, serine-rich 2-like |
4520 |
0.28 |
| chr3_27710920_27711171 | 0.08 |
Fndc3b |
fibronectin type III domain containing 3B |
194 |
0.97 |
| chr13_35817740_35817891 | 0.07 |
Gm48706 |
predicted gene, 48706 |
19065 |
0.16 |
| chr15_62934546_62934877 | 0.07 |
Tsg101-ps |
tumor susceptibility gene 101, pseudogene |
45937 |
0.19 |
| chr17_47953060_47953435 | 0.07 |
Gm5228 |
predicted gene 5228 |
17157 |
0.13 |
| chr6_114875640_114875809 | 0.07 |
Vgll4 |
vestigial like family member 4 |
617 |
0.75 |
| chr1_177729767_177729918 | 0.07 |
1700016C15Rik |
RIKEN cDNA 1700016C15 gene |
28 |
0.98 |
| chr12_109038321_109038660 | 0.06 |
Begain |
brain-enriched guanylate kinase-associated |
1403 |
0.37 |
| chr15_76084268_76084515 | 0.06 |
Gm48952 |
predicted gene, 48952 |
627 |
0.46 |
| chr9_29514897_29515079 | 0.06 |
Gm15521 |
predicted gene 15521 |
77522 |
0.12 |
| chr5_32611305_32611927 | 0.06 |
Gm10461 |
predicted gene 10461 |
189 |
0.6 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.3 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0003150 | muscular septum morphogenesis(GO:0003150) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:1903261 | regulation of serine phosphorylation of STAT3 protein(GO:1903261) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.2 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 | 0.1 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 0.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.0 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 0.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.4 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |