Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
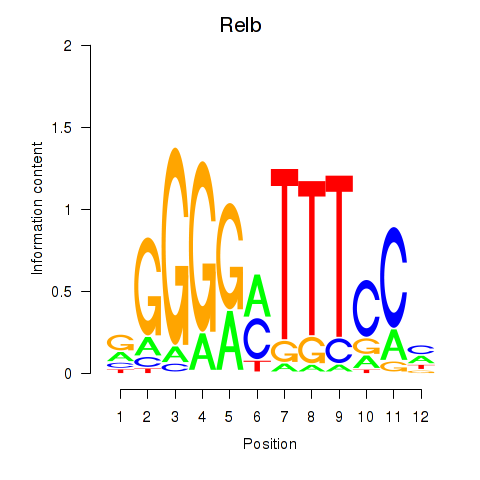
Results for Relb
Z-value: 0.76
Transcription factors associated with Relb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Relb
|
ENSMUSG00000002983.10 | avian reticuloendotheliosis viral (v-rel) oncogene related B |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_19629423_19629638 | Relb | 92 | 0.928645 | -0.57 | 5.7e-06 | Click! |
| chr7_19619101_19619365 | Relb | 9201 | 0.084886 | -0.38 | 4.4e-03 | Click! |
| chr7_19632370_19632657 | Relb | 3075 | 0.113409 | 0.36 | 7.1e-03 | Click! |
| chr7_19628297_19628448 | Relb | 62 | 0.938815 | -0.33 | 1.4e-02 | Click! |
| chr7_19628520_19629349 | Relb | 150 | 0.900024 | -0.29 | 3.2e-02 | Click! |
Activity of the Relb motif across conditions
Conditions sorted by the z-value of the Relb motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_135721881_135722214 | 3.02 |
Mki67 |
antigen identified by monoclonal antibody Ki 67 |
5686 |
0.18 |
| chr11_57952722_57952903 | 2.91 |
Gm12245 |
predicted gene 12245 |
3727 |
0.2 |
| chr6_124646261_124646412 | 2.51 |
C1s2 |
complement component 1, s subcomponent 2 |
10251 |
0.09 |
| chr11_23770360_23771742 | 2.36 |
Rel |
reticuloendotheliosis oncogene |
81 |
0.97 |
| chr7_141628110_141628282 | 2.35 |
Mir7063 |
microRNA 7063 |
7495 |
0.13 |
| chr19_29406708_29406859 | 2.14 |
Pdcd1lg2 |
programmed cell death 1 ligand 2 |
4136 |
0.18 |
| chr4_152092303_152092489 | 1.97 |
Plekhg5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
4323 |
0.14 |
| chrX_150565075_150565302 | 1.85 |
Alas2 |
aminolevulinic acid synthase 2, erythroid |
1202 |
0.4 |
| chr2_122138001_122138389 | 1.83 |
B2m |
beta-2 microglobulin |
9491 |
0.12 |
| chr17_84729001_84729167 | 1.71 |
Lrpprc |
leucine-rich PPR-motif containing |
2100 |
0.28 |
| chr19_41347313_41347632 | 1.66 |
Pik3ap1 |
phosphoinositide-3-kinase adaptor protein 1 |
37624 |
0.17 |
| chr15_84168741_84169197 | 1.64 |
Mir6392 |
microRNA 6392 |
699 |
0.44 |
| chr8_46739090_46739964 | 1.61 |
Gm16675 |
predicted gene, 16675 |
12 |
0.73 |
| chr11_32220828_32222435 | 1.60 |
Rhbdf1 |
rhomboid 5 homolog 1 |
613 |
0.58 |
| chr12_91696294_91696631 | 1.54 |
Gm8378 |
predicted gene 8378 |
10349 |
0.16 |
| chr4_120665617_120665768 | 1.47 |
Cited4 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
880 |
0.53 |
| chr4_150787186_150787387 | 1.41 |
Gm13049 |
predicted gene 13049 |
38447 |
0.13 |
| chr3_153792067_153792514 | 1.39 |
5730460C07Rik |
RIKEN cDNA 5730460C07 gene |
203 |
0.9 |
| chr2_28599202_28599576 | 1.38 |
Gm22675 |
predicted gene, 22675 |
4262 |
0.12 |
| chr13_101552934_101553307 | 1.36 |
Gm19010 |
predicted gene, 19010 |
4711 |
0.18 |
| chr11_37412938_37413250 | 1.35 |
Tenm2 |
teneurin transmembrane protein 2 |
177130 |
0.03 |
| chr6_120619174_120619360 | 1.33 |
Gm44124 |
predicted gene, 44124 |
39091 |
0.11 |
| chr9_70934534_70934685 | 1.32 |
Lipc |
lipase, hepatic |
4 |
0.98 |
| chr11_109584714_109584865 | 1.32 |
Wipi1 |
WD repeat domain, phosphoinositide interacting 1 |
26643 |
0.13 |
| chr8_104886596_104886898 | 1.31 |
Gm8798 |
predicted gene 8798 |
115 |
0.93 |
| chr8_104867227_104867534 | 1.30 |
Ces2d-ps |
carboxylesterase 2D, pseudogene |
108 |
0.93 |
| chr17_74331488_74331639 | 1.29 |
Gm9351 |
predicted gene 9351 |
966 |
0.36 |
| chr17_75391941_75392106 | 1.25 |
Ltbp1 |
latent transforming growth factor beta binding protein 1 |
7813 |
0.27 |
| chr3_146379986_146380483 | 1.25 |
Gm10636 |
predicted gene 10636 |
1290 |
0.35 |
| chr5_140583084_140583332 | 1.21 |
Grifin |
galectin-related inter-fiber protein |
18137 |
0.11 |
| chr13_59639448_59639751 | 1.20 |
Naa35 |
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
6874 |
0.12 |
| chr1_177484652_177484803 | 1.19 |
Gm37306 |
predicted gene, 37306 |
17349 |
0.16 |
| chr1_98094869_98096038 | 1.13 |
Pam |
peptidylglycine alpha-amidating monooxygenase |
168 |
0.95 |
| chr5_88699513_88699664 | 1.10 |
Mob1b |
MOB kinase activator 1B |
21267 |
0.13 |
| chr12_118521494_118521645 | 1.09 |
D230030E09Rik |
Riken cDNA D230030E09 gene |
8624 |
0.23 |
| chr1_160044783_160045062 | 1.09 |
4930523C07Rik |
RIKEN cDNA 4930523C07 gene |
475 |
0.52 |
| chr4_108317802_108317953 | 1.08 |
Gm12740 |
predicted gene 12740 |
4461 |
0.14 |
| chr11_102375203_102375512 | 1.08 |
Bloodlinc |
Bloodlinc, erythroid developmental long intergenic non-protein coding transcript |
1663 |
0.21 |
| chr8_89524816_89524967 | 1.07 |
Gm26331 |
predicted gene, 26331 |
64650 |
0.15 |
| chr11_116023790_116023980 | 1.06 |
H3f3b |
H3.3 histone B |
588 |
0.58 |
| chr8_70863265_70863898 | 1.04 |
Kcnn1 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
323 |
0.53 |
| chr3_83097912_83098063 | 1.03 |
Plrg1 |
pleiotropic regulator 1 |
29014 |
0.13 |
| chr10_115780298_115780520 | 1.02 |
Tspan8 |
tetraspanin 8 |
36423 |
0.2 |
| chr11_34783136_34783287 | 1.01 |
Dock2 |
dedicator of cyto-kinesis 2 |
672 |
0.75 |
| chr5_115626548_115626814 | 1.01 |
1110006O24Rik |
RIKEN cDNA 1110006O24 gene |
5135 |
0.11 |
| chr10_62215728_62215879 | 0.99 |
Tspan15 |
tetraspanin 15 |
14026 |
0.15 |
| chr7_141103278_141103452 | 0.97 |
Ano9 |
anoctamin 9 |
689 |
0.42 |
| chr19_48310424_48310575 | 0.96 |
Gm23857 |
predicted gene, 23857 |
69854 |
0.12 |
| chr11_113467252_113467828 | 0.95 |
Gm16487 |
predicted gene 16487 |
437 |
0.89 |
| chr1_85114233_85115011 | 0.94 |
Gm16038 |
predicted gene 16038 |
697 |
0.48 |
| chr13_64179369_64179540 | 0.94 |
Habp4 |
hyaluronic acid binding protein 4 |
5049 |
0.13 |
| chr1_136859565_136859716 | 0.92 |
Gm17781 |
predicted gene, 17781 |
60681 |
0.09 |
| chr7_121348775_121349004 | 0.91 |
Hs3st2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
42970 |
0.13 |
| chr19_17491820_17492164 | 0.91 |
Rfk |
riboflavin kinase |
94555 |
0.08 |
| chr11_78163296_78164397 | 0.91 |
Traf4 |
TNF receptor associated factor 4 |
1670 |
0.14 |
| chr19_7017655_7018362 | 0.91 |
Fermt3 |
fermitin family member 3 |
1337 |
0.22 |
| chr2_50964143_50964317 | 0.90 |
Gm13498 |
predicted gene 13498 |
54546 |
0.16 |
| chr6_51320650_51320992 | 0.90 |
Gm32479 |
predicted gene, 32479 |
32966 |
0.15 |
| chr7_90060924_90061075 | 0.90 |
Gm44861 |
predicted gene 44861 |
18302 |
0.12 |
| chr3_5210842_5211599 | 0.90 |
Gm10748 |
predicted gene 10748 |
1618 |
0.38 |
| chr1_130832611_130832799 | 0.90 |
Pigr |
polymeric immunoglobulin receptor |
3695 |
0.15 |
| chr8_53368812_53368963 | 0.88 |
Gm45554 |
predicted gene 45554 |
67213 |
0.13 |
| chr1_176813990_176814449 | 0.87 |
Cep170 |
centrosomal protein 170 |
152 |
0.78 |
| chr1_86312319_86312630 | 0.87 |
Gm28626 |
predicted gene 28626 |
8415 |
0.08 |
| chr9_91193771_91193922 | 0.87 |
Gm29602 |
predicted gene 29602 |
22169 |
0.17 |
| chr8_81527747_81528046 | 0.87 |
4930579O11Rik |
RIKEN cDNA 4930579O11 gene |
139176 |
0.05 |
| chr1_138559713_138559887 | 0.86 |
Gm37080 |
predicted gene, 37080 |
5092 |
0.19 |
| chr1_85243802_85244548 | 0.85 |
C130026I21Rik |
RIKEN cDNA C130026I21 gene |
2222 |
0.19 |
| chr17_65557305_65557480 | 0.85 |
Gm49866 |
predicted gene, 49866 |
3384 |
0.19 |
| chr16_9680012_9680359 | 0.84 |
Grin2a |
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
117148 |
0.07 |
| chr7_103870271_103870435 | 0.83 |
Olfr66 |
olfactory receptor 66 |
11888 |
0.06 |
| chr1_105784247_105784562 | 0.83 |
Tnfrsf11a |
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
3654 |
0.21 |
| chr4_123411292_123412523 | 0.83 |
Macf1 |
microtubule-actin crosslinking factor 1 |
321 |
0.88 |
| chr2_153483459_153484019 | 0.83 |
Nol4l |
nucleolar protein 4-like |
87 |
0.97 |
| chr10_26949652_26949996 | 0.82 |
Gm48084 |
predicted gene, 48084 |
84 |
0.97 |
| chr10_16600909_16601254 | 0.82 |
Gm47729 |
predicted gene, 47729 |
16813 |
0.28 |
| chr5_123180971_123181122 | 0.81 |
Hpd |
4-hydroxyphenylpyruvic acid dioxygenase |
1021 |
0.33 |
| chr9_64591461_64591626 | 0.79 |
Megf11 |
multiple EGF-like-domains 11 |
44644 |
0.16 |
| chr9_64049464_64049751 | 0.79 |
Gm25606 |
predicted gene, 25606 |
1111 |
0.4 |
| chr9_50693044_50693195 | 0.78 |
Dixdc1 |
DIX domain containing 1 |
680 |
0.59 |
| chr2_133400784_133400978 | 0.78 |
A430048G15Rik |
RIKEN cDNA A430048G15 gene |
31214 |
0.18 |
| chr9_64566375_64566728 | 0.78 |
Megf11 |
multiple EGF-like-domains 11 |
19652 |
0.24 |
| chr13_81663841_81663992 | 0.77 |
Lysmd3 |
LysM, putative peptidoglycan-binding, domain containing 3 |
1278 |
0.42 |
| chr1_161968669_161969306 | 0.77 |
4930558K02Rik |
RIKEN cDNA 4930558K02 gene |
161 |
0.39 |
| chr16_38406899_38407050 | 0.77 |
Pla1a |
phospholipase A1 member A |
10820 |
0.12 |
| chr11_98459235_98459494 | 0.76 |
Grb7 |
growth factor receptor bound protein 7 |
5575 |
0.1 |
| chr17_87365323_87365495 | 0.75 |
0610012D04Rik |
RIKEN cDNA 0610012D04 gene |
947 |
0.48 |
| chr9_124312159_124312527 | 0.75 |
2010315B03Rik |
RIKEN cDNA 2010315B03 gene |
299 |
0.86 |
| chr5_129489887_129490038 | 0.75 |
Gm40332 |
predicted gene, 40332 |
11039 |
0.18 |
| chr8_117718550_117718744 | 0.74 |
Hsd17b2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
2289 |
0.22 |
| chr10_37440176_37440344 | 0.74 |
Gm48172 |
predicted gene, 48172 |
12420 |
0.29 |
| chr15_31571122_31571273 | 0.74 |
Cmbl |
carboxymethylenebutenolidase-like (Pseudomonas) |
908 |
0.48 |
| chr1_75443276_75444022 | 0.73 |
Gmppa |
GDP-mannose pyrophosphorylase A |
1425 |
0.23 |
| chr18_4862423_4862746 | 0.73 |
Gm10556 |
predicted gene 10556 |
50098 |
0.15 |
| chr3_18206079_18206269 | 0.72 |
Gm23686 |
predicted gene, 23686 |
28549 |
0.18 |
| chr7_4748180_4748350 | 0.72 |
Kmt5c |
lysine methyltransferase 5C |
2457 |
0.11 |
| chr9_123386502_123386682 | 0.72 |
Lars2 |
leucyl-tRNA synthetase, mitochondrial |
8856 |
0.2 |
| chr7_19867840_19867991 | 0.72 |
Gm44659 |
predicted gene 44659 |
3523 |
0.09 |
| chr10_121445896_121446136 | 0.71 |
Rassf3 |
Ras association (RalGDS/AF-6) domain family member 3 |
16197 |
0.12 |
| chr19_41699158_41699309 | 0.70 |
Slit1 |
slit guidance ligand 1 |
44253 |
0.14 |
| chr3_152113520_152113951 | 0.70 |
Gipc2 |
GIPC PDZ domain containing family, member 2 |
5213 |
0.15 |
| chr10_120956876_120957207 | 0.70 |
Gm23632 |
predicted gene, 23632 |
17384 |
0.12 |
| chr12_76558245_76558525 | 0.70 |
Plekhg3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
3853 |
0.17 |
| chr2_24408191_24408394 | 0.69 |
Psd4 |
pleckstrin and Sec7 domain containing 4 |
2663 |
0.21 |
| chr2_103970312_103971317 | 0.69 |
Lmo2 |
LIM domain only 2 |
365 |
0.83 |
| chr7_46724003_46724164 | 0.69 |
Saa3 |
serum amyloid A 3 |
8383 |
0.09 |
| chr7_96944132_96944283 | 0.69 |
C230038L03Rik |
RIKEN cDNA C230038L03 gene |
2827 |
0.24 |
| chr1_151177160_151177572 | 0.68 |
Gm47985 |
predicted gene, 47985 |
5171 |
0.12 |
| chr1_85581692_85582254 | 0.68 |
Sp110 |
Sp110 nuclear body protein |
2305 |
0.16 |
| chr6_136938860_136939011 | 0.67 |
Arhgdib |
Rho, GDP dissociation inhibitor (GDI) beta |
656 |
0.62 |
| chr10_128590143_128590705 | 0.67 |
Erbb3 |
erb-b2 receptor tyrosine kinase 3 |
772 |
0.35 |
| chr8_123051510_123051721 | 0.67 |
2810013P06Rik |
RIKEN cDNA 2810013P06 gene |
9149 |
0.1 |
| chr6_41556535_41556686 | 0.66 |
Trbj2-6 |
T cell receptor beta joining 2-6 |
13012 |
0.09 |
| chr15_96709352_96710351 | 0.65 |
Gm38144 |
predicted gene, 38144 |
9678 |
0.18 |
| chr6_57513773_57513924 | 0.65 |
Ppm1k |
protein phosphatase 1K (PP2C domain containing) |
1759 |
0.29 |
| chr5_15618415_15618598 | 0.65 |
Speer4d |
spermatogenesis associated glutamate (E)-rich protein 4D |
558 |
0.71 |
| chr3_90560096_90560247 | 0.65 |
S100a2 |
S100 calcium binding protein A2 |
192 |
0.73 |
| chr1_85092888_85093459 | 0.64 |
Gm10553 |
predicted gene 10553 |
6339 |
0.1 |
| chr19_4554348_4554539 | 0.64 |
Pcx |
pyruvate carboxylase |
4367 |
0.16 |
| chr9_34965089_34965274 | 0.64 |
Gm23702 |
predicted gene, 23702 |
5162 |
0.22 |
| chr18_36560129_36560675 | 0.64 |
Ankhd1 |
ankyrin repeat and KH domain containing 1 |
209 |
0.91 |
| chr19_38164769_38164937 | 0.63 |
Pde6c |
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
31907 |
0.13 |
| chr5_90504957_90505176 | 0.63 |
Afp |
alpha fetoprotein |
2365 |
0.21 |
| chr4_104986072_104986223 | 0.63 |
Gm12721 |
predicted gene 12721 |
16018 |
0.24 |
| chr3_116423430_116424024 | 0.63 |
Cdc14a |
CDC14 cell division cycle 14A |
168 |
0.94 |
| chrX_10877865_10878215 | 0.62 |
Gm14473 |
predicted gene 14473 |
12407 |
0.26 |
| chr4_106791303_106791454 | 0.62 |
Acot11 |
acyl-CoA thioesterase 11 |
8412 |
0.16 |
| chr6_3384964_3385136 | 0.62 |
Samd9l |
sterile alpha motif domain containing 9-like |
44 |
0.97 |
| chr5_135118609_135119147 | 0.61 |
Gm43500 |
predicted gene 43500 |
4415 |
0.12 |
| chr16_56029023_56029537 | 0.61 |
Pcnp |
PEST proteolytic signal containing nuclear protein |
396 |
0.74 |
| chr5_92359200_92359362 | 0.61 |
Cxcl11 |
chemokine (C-X-C motif) ligand 11 |
3996 |
0.12 |
| chr5_14979304_14979699 | 0.60 |
Gm10354 |
predicted gene 10354 |
566 |
0.78 |
| chr14_79390171_79390957 | 0.60 |
Naa16 |
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
126 |
0.95 |
| chr11_53483659_53483860 | 0.60 |
Sowaha |
sosondowah ankyrin repeat domain family member A |
3485 |
0.1 |
| chr9_15534236_15534387 | 0.60 |
Smco4 |
single-pass membrane protein with coiled-coil domains 4 |
13452 |
0.17 |
| chr4_43957119_43958388 | 0.59 |
Glipr2 |
GLI pathogenesis-related 2 |
61 |
0.96 |
| chr2_151743607_151743758 | 0.59 |
Psmf1 |
proteasome (prosome, macropain) inhibitor subunit 1 |
504 |
0.7 |
| chr5_22009261_22009412 | 0.59 |
Reln |
reelin |
928 |
0.6 |
| chr4_125122466_125122637 | 0.59 |
Zc3h12a |
zinc finger CCCH type containing 12A |
31 |
0.97 |
| chr8_94171665_94171816 | 0.59 |
Mt2 |
metallothionein 2 |
924 |
0.35 |
| chr3_98283507_98283658 | 0.59 |
Hmgcs2 |
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 |
3147 |
0.18 |
| chr17_64200169_64200637 | 0.58 |
Pja2 |
praja ring finger ubiquitin ligase 2 |
112628 |
0.07 |
| chr2_130638104_130638350 | 0.58 |
Lzts3 |
leucine zipper, putative tumor suppressor family member 3 |
132 |
0.92 |
| chr3_24605465_24605826 | 0.58 |
Gm24704 |
predicted gene, 24704 |
43571 |
0.21 |
| chr9_40465704_40465985 | 0.57 |
Gramd1b |
GRAM domain containing 1B |
280 |
0.89 |
| chr16_30063252_30064537 | 0.57 |
Hes1 |
hes family bHLH transcription factor 1 |
490 |
0.76 |
| chr17_24204910_24205196 | 0.57 |
Tbc1d24 |
TBC1 domain family, member 24 |
429 |
0.64 |
| chr8_84934869_84935487 | 0.57 |
Mast1 |
microtubule associated serine/threonine kinase 1 |
2166 |
0.11 |
| chr9_42461386_42461564 | 0.57 |
Tbcel |
tubulin folding cofactor E-like |
14 |
0.98 |
| chr15_82183804_82184036 | 0.56 |
Gm49502 |
predicted gene, 49502 |
1596 |
0.21 |
| chr2_133373650_133373819 | 0.56 |
Gm27449 |
predicted gene, 27449 |
42810 |
0.16 |
| chr10_91859325_91859494 | 0.56 |
Gm31592 |
predicted gene, 31592 |
29420 |
0.24 |
| chr5_135115423_135115665 | 0.56 |
Gm43500 |
predicted gene 43500 |
7749 |
0.1 |
| chr11_88420075_88420250 | 0.56 |
Gm11510 |
predicted gene 11510 |
13352 |
0.2 |
| chr8_120494857_120495334 | 0.55 |
Gse1 |
genetic suppressor element 1, coiled-coil protein |
6648 |
0.15 |
| chr4_125124451_125124813 | 0.55 |
Zc3h12a |
zinc finger CCCH type containing 12A |
2112 |
0.25 |
| chr5_14938987_14939138 | 0.54 |
Speer4e |
spermatogenesis associated glutamate (E)-rich protein 4e |
633 |
0.73 |
| chr4_46040327_46040637 | 0.54 |
Tmod1 |
tropomodulin 1 |
1273 |
0.46 |
| chr6_136497922_136498073 | 0.54 |
Atf7ip |
activating transcription factor 7 interacting protein |
8170 |
0.12 |
| chr7_81656674_81656984 | 0.54 |
Gm26149 |
predicted gene, 26149 |
12899 |
0.13 |
| chr5_112342677_112343550 | 0.54 |
Hps4 |
HPS4, biogenesis of lysosomal organelles complex 3 subunit 2 |
13 |
0.54 |
| chr2_33431513_33431719 | 0.54 |
Zbtb34 |
zinc finger and BTB domain containing 34 |
292 |
0.88 |
| chr9_5308573_5308892 | 0.54 |
Casp4 |
caspase 4, apoptosis-related cysteine peptidase |
96 |
0.97 |
| chr18_84886022_84886173 | 0.54 |
Cyb5a |
cytochrome b5 type A (microsomal) |
8496 |
0.17 |
| chr6_122874335_122875052 | 0.54 |
Necap1 |
NECAP endocytosis associated 1 |
112 |
0.94 |
| chr4_129140170_129140321 | 0.53 |
Fndc5 |
fibronectin type III domain containing 5 |
3246 |
0.15 |
| chr5_15476867_15477289 | 0.53 |
Gm21149 |
predicted gene, 21149 |
555 |
0.74 |
| chr2_172727961_172728141 | 0.53 |
Gm22773 |
predicted gene, 22773 |
136290 |
0.04 |
| chr4_129242022_129242244 | 0.53 |
C77080 |
expressed sequence C77080 |
2959 |
0.17 |
| chr4_14865526_14865698 | 0.53 |
Pip4p2 |
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
1536 |
0.43 |
| chr4_144887592_144887743 | 0.53 |
Dhrs3 |
dehydrogenase/reductase (SDR family) member 3 |
5160 |
0.24 |
| chr9_96979442_96979784 | 0.53 |
Spsb4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
4582 |
0.19 |
| chr12_24633832_24634168 | 0.53 |
Gm6969 |
predicted pseudogene 6969 |
5010 |
0.17 |
| chr18_4921251_4922052 | 0.53 |
Svil |
supervillin |
75 |
0.98 |
| chr3_104938728_104938879 | 0.53 |
Gm43846 |
predicted gene 43846 |
19570 |
0.12 |
| chr4_154414883_154415251 | 0.52 |
Prdm16 |
PR domain containing 16 |
66610 |
0.1 |
| chr9_64086915_64087080 | 0.52 |
Scarletltr |
Scarletltr, erythroid developmental long intergenic non-protein coding transcript |
6236 |
0.13 |
| chr7_100493605_100494145 | 0.52 |
Ucp2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
80 |
0.94 |
| chr2_35610268_35610691 | 0.52 |
Dab2ip |
disabled 2 interacting protein |
11502 |
0.2 |
| chr10_67003002_67003552 | 0.52 |
Gm31763 |
predicted gene, 31763 |
1743 |
0.37 |
| chr10_108434084_108434235 | 0.52 |
Gm36283 |
predicted gene, 36283 |
2506 |
0.28 |
| chr11_75637667_75638005 | 0.51 |
Inpp5k |
inositol polyphosphate 5-phosphatase K |
1301 |
0.31 |
| chr17_83956064_83956273 | 0.51 |
Gm35229 |
predicted gene, 35229 |
80 |
0.95 |
| chr18_47342335_47342491 | 0.51 |
Sema6a |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
9065 |
0.23 |
| chr13_80382232_80382383 | 0.51 |
Gm46388 |
predicted gene, 46388 |
110702 |
0.07 |
| chr2_158302879_158303150 | 0.50 |
Lbp |
lipopolysaccharide binding protein |
3479 |
0.15 |
| chr7_28596265_28596676 | 0.50 |
Pak4 |
p21 (RAC1) activated kinase 4 |
1616 |
0.18 |
| chr17_27123920_27124092 | 0.50 |
Uqcc2 |
ubiquinol-cytochrome c reductase complex assembly factor 2 |
9617 |
0.09 |
| chr2_20689498_20689881 | 0.50 |
Gm13362 |
predicted gene 13362 |
23545 |
0.21 |
| chr1_168287303_168287456 | 0.50 |
Gm37524 |
predicted gene, 37524 |
50292 |
0.16 |
| chr6_84327421_84327611 | 0.50 |
Gm10445 |
predicted gene 10445 |
96322 |
0.07 |
| chr6_91121562_91121713 | 0.50 |
Gm44278 |
predicted gene, 44278 |
702 |
0.61 |
| chr9_27308659_27309037 | 0.50 |
Igsf9b |
immunoglobulin superfamily, member 9B |
9620 |
0.19 |
| chr15_102460993_102461420 | 0.50 |
Prr13 |
proline rich 13 |
1123 |
0.3 |
| chr11_83578517_83578693 | 0.50 |
Ccl9 |
chemokine (C-C motif) ligand 9 |
31 |
0.95 |
| chr15_59706829_59706980 | 0.49 |
Gm20150 |
predicted gene, 20150 |
34791 |
0.17 |
| chr7_56765957_56766108 | 0.49 |
Gm44979 |
predicted gene 44979 |
17758 |
0.2 |
| chr10_19014594_19015624 | 0.49 |
Tnfaip3 |
tumor necrosis factor, alpha-induced protein 3 |
267 |
0.93 |
| chr5_139300674_139300825 | 0.49 |
Adap1 |
ArfGAP with dual PH domains 1 |
2401 |
0.22 |
| chr18_46198704_46199602 | 0.48 |
1700018A14Rik |
RIKEN cDNA 1700018A14 gene |
132 |
0.75 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.3 | 0.8 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.3 | 0.8 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.2 | 0.4 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.2 | 1.3 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.2 | 0.6 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.2 | 0.5 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.2 | 0.5 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.1 | 0.4 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.6 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 0.4 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.1 | 0.5 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.5 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.3 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 1.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.4 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.6 | GO:0051256 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.5 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 0.4 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.3 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.3 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.1 | 0.3 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.1 | 0.2 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.1 | 0.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.2 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.1 | 0.2 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.1 | 0.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.2 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 | 0.2 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.3 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.2 | GO:0002017 | regulation of blood volume by renal aldosterone(GO:0002017) |
| 0.1 | 0.2 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.1 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:0010535 | positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.1 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.2 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.8 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.2 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 0.3 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.1 | 0.3 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.1 | 0.3 | GO:0034756 | regulation of iron ion transport(GO:0034756) |
| 0.1 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.5 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 0.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.2 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.2 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 | 0.2 | GO:0045869 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) |
| 0.1 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.3 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.2 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.0 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0072235 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0042504 | tyrosine phosphorylation of Stat4 protein(GO:0042504) regulation of tyrosine phosphorylation of Stat4 protein(GO:0042519) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0060618 | nipple development(GO:0060618) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0009757 | carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.3 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.0 | 0.1 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.2 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.2 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.0 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.0 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0098910 | regulation of atrial cardiac muscle cell action potential(GO:0098910) |
| 0.0 | 0.0 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.0 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.0 | 0.3 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0015817 | histidine transport(GO:0015817) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0001907 | killing by symbiont of host cells(GO:0001907) disruption by symbiont of host cell(GO:0044004) |
| 0.0 | 0.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.0 | GO:0038086 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.0 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.3 | GO:0010714 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.0 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.0 | 0.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.0 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.0 | GO:0060375 | regulation of mast cell differentiation(GO:0060375) |
| 0.0 | 0.0 | GO:0061046 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 0.0 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.0 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.0 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.0 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900164) |
| 0.0 | 0.1 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.1 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.0 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.0 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.0 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:2000615 | regulation of histone H3-K9 acetylation(GO:2000615) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.0 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.0 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.0 | 0.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0034116 | positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.0 | 0.0 | GO:2000416 | regulation of eosinophil migration(GO:2000416) positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.0 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.0 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0061450 | trophoblast cell migration(GO:0061450) |
| 0.0 | 0.0 | GO:1901628 | positive regulation of postsynaptic membrane organization(GO:1901628) positive regulation of receptor clustering(GO:1903911) regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.0 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.2 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 0.1 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.3 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.0 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0071549 | response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.0 | GO:0050427 | 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.2 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.5 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.4 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.1 | 0.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.2 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.2 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.0 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 1.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 1.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 1.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.0 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.0 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.0 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.0 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.3 | 1.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 0.6 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.6 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.5 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.4 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 2.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 0.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.8 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.2 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.2 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 1.3 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0044466 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0034889 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0015186 | L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0003933 | GTP cyclohydrolase activity(GO:0003933) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0016443 | bidentate ribonuclease III activity(GO:0016443) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.3 | GO:0000252 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0003905 | alkylbase DNA N-glycosylase activity(GO:0003905) DNA-3-methylbase glycosylase activity(GO:0043733) |
| 0.0 | 0.0 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.0 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.0 | 0.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 0.7 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 2.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.0 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.5 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |