Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
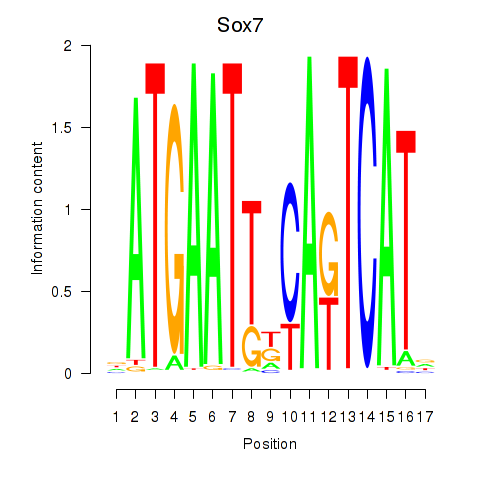
Results for Sox7
Z-value: 1.80
Transcription factors associated with Sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox7
|
ENSMUSG00000063060.5 | SRY (sex determining region Y)-box 7 |
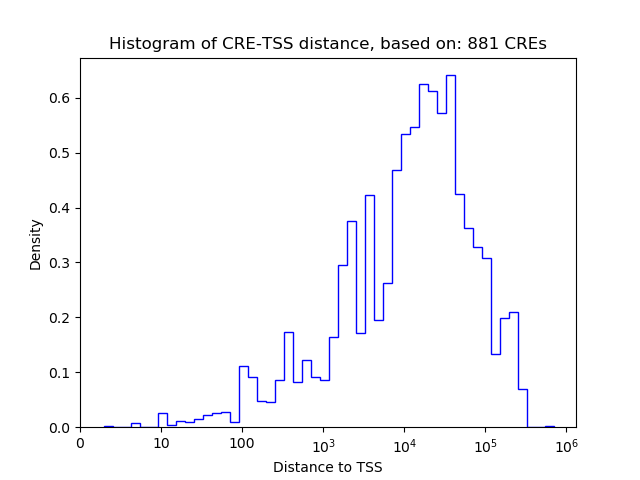
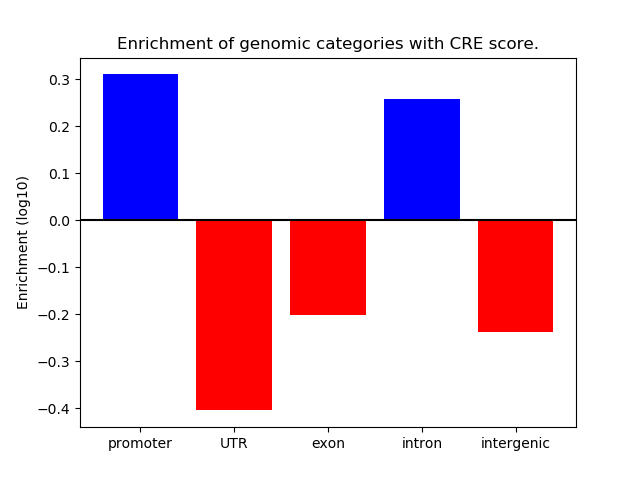
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_63957850_63958017 | Sox7 | 14260 | 0.157862 | -0.45 | 6.6e-04 | Click! |
| chr14_63958056_63958207 | Sox7 | 14458 | 0.157398 | -0.38 | 4.1e-03 | Click! |
| chr14_63943484_63943756 | Sox7 | 53 | 0.973669 | -0.26 | 5.9e-02 | Click! |
| chr14_63962821_63963010 | Sox7 | 19242 | 0.146143 | 0.06 | 6.5e-01 | Click! |
| chr14_63943854_63944005 | Sox7 | 256 | 0.915038 | 0.00 | 9.8e-01 | Click! |
Activity of the Sox7 motif across conditions
Conditions sorted by the z-value of the Sox7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
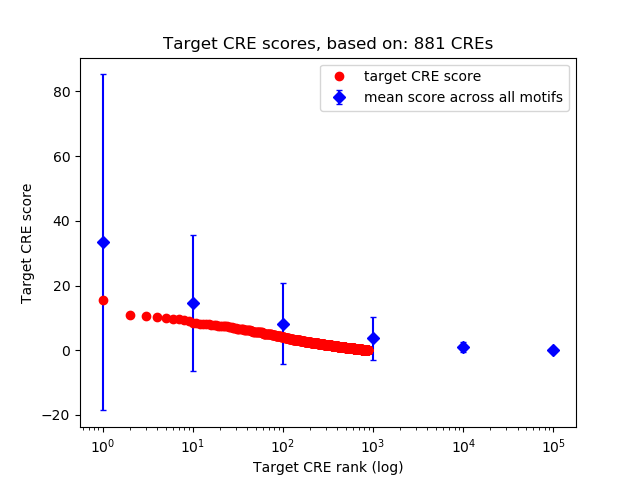
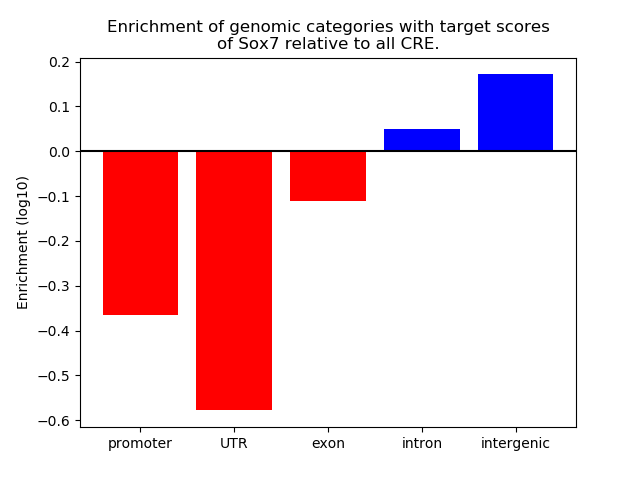
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_112695608_112695943 | 15.64 |
Gm18883 |
predicted gene, 18883 |
3904 |
0.16 |
| chr2_70168731_70168924 | 10.86 |
Myo3b |
myosin IIIB |
72529 |
0.11 |
| chr10_23674681_23675112 | 10.73 |
4930520K02Rik |
RIKEN cDNA 4930520K02 gene |
52748 |
0.11 |
| chr4_116016940_116018214 | 10.39 |
Faah |
fatty acid amide hydrolase |
98 |
0.95 |
| chr6_114289060_114289436 | 10.07 |
Slc6a1 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 1 |
6458 |
0.27 |
| chr4_109763223_109763401 | 9.62 |
Gm12808 |
predicted gene 12808 |
8495 |
0.24 |
| chr18_45008774_45008950 | 9.61 |
Gm31706 |
predicted gene, 31706 |
36000 |
0.15 |
| chr1_30751228_30751385 | 9.46 |
Gm6473 |
predicted gene 6473 |
18381 |
0.17 |
| chr3_11024565_11024725 | 9.11 |
Gm21631 |
predicted gene, 21631 |
15042 |
0.2 |
| chr12_29723997_29724347 | 8.56 |
C630031E19Rik |
RIKEN cDNA C630031E19 gene |
37727 |
0.21 |
| chr3_87098356_87098639 | 8.51 |
Gm24472 |
predicted gene, 24472 |
31604 |
0.12 |
| chr11_44977922_44978106 | 8.25 |
Ebf1 |
early B cell factor 1 |
65876 |
0.13 |
| chr13_28295576_28295772 | 8.14 |
Gm47172 |
predicted gene, 47172 |
10105 |
0.23 |
| chr10_16052455_16052829 | 8.10 |
C330004P14Rik |
RIKEN cDNA C330004P14 gene |
260865 |
0.02 |
| chr7_78578810_78579126 | 8.09 |
Gm9885 |
predicted gene 9885 |
138 |
0.68 |
| chr3_39857262_39857458 | 7.78 |
Gm42785 |
predicted gene 42785 |
33783 |
0.17 |
| chr13_97253425_97254036 | 7.74 |
Enc1 |
ectodermal-neural cortex 1 |
12625 |
0.16 |
| chr12_51002470_51002650 | 7.69 |
Gm40421 |
predicted gene, 40421 |
2313 |
0.29 |
| chr10_120377883_120378034 | 7.63 |
9230105E05Rik |
RIKEN cDNA 9230105E05 gene |
11565 |
0.16 |
| chr6_36776057_36776246 | 7.60 |
Ptn |
pleiotrophin |
34028 |
0.2 |
| chr12_86680804_86681001 | 7.54 |
Vash1 |
vasohibin 1 |
2202 |
0.24 |
| chr7_25006131_25006556 | 7.41 |
Atp1a3 |
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
385 |
0.75 |
| chr13_15624842_15625048 | 7.41 |
Gm48343 |
predicted gene, 48343 |
24990 |
0.22 |
| chr6_7601514_7601665 | 7.35 |
Tac1 |
tachykinin 1 |
39221 |
0.14 |
| chr9_47985729_47986075 | 7.30 |
Gm47236 |
predicted gene, 47236 |
14896 |
0.18 |
| chr13_28690458_28690659 | 7.12 |
Mir6368 |
microRNA 6368 |
20315 |
0.25 |
| chr7_60450080_60450452 | 7.10 |
Gm30196 |
predicted gene, 30196 |
157062 |
0.03 |
| chr10_18468496_18468770 | 6.99 |
Nhsl1 |
NHS-like 1 |
1255 |
0.55 |
| chr6_54573357_54573532 | 6.90 |
Scrn1 |
secernin 1 |
6955 |
0.17 |
| chr1_172028700_172028921 | 6.75 |
Vangl2 |
VANGL planar cell polarity 2 |
366 |
0.81 |
| chr4_47772365_47772516 | 6.61 |
Gm22670 |
predicted gene, 22670 |
148315 |
0.04 |
| chr7_143385947_143386098 | 6.61 |
4933417O13Rik |
RIKEN cDNA 4933417O13 gene |
21397 |
0.13 |
| chr7_120157803_120157967 | 6.58 |
4930505K13Rik |
RIKEN cDNA 4930505K13 gene |
1770 |
0.26 |
| chr1_168908922_168909127 | 6.56 |
Mir6354 |
microRNA 6354 |
110065 |
0.07 |
| chr8_100939492_100939914 | 6.46 |
Gm39232 |
predicted gene, 39232 |
213348 |
0.02 |
| chr2_123349959_123350330 | 6.43 |
Gm13988 |
predicted gene 13988 |
76220 |
0.13 |
| chr8_16013714_16013865 | 6.35 |
Csmd1 |
CUB and Sushi multiple domains 1 |
28977 |
0.24 |
| chr17_55406170_55406321 | 6.35 |
Gm49921 |
predicted gene, 49921 |
17538 |
0.21 |
| chr6_78245012_78245212 | 6.32 |
Gm5576 |
predicted pseudogene 5576 |
29234 |
0.21 |
| chr15_34824497_34824736 | 6.30 |
Gm48932 |
predicted gene, 48932 |
1813 |
0.39 |
| chr6_103513728_103513971 | 6.25 |
Chl1 |
cell adhesion molecule L1-like |
2519 |
0.25 |
| chr9_92173926_92174156 | 6.18 |
Plscr5 |
phospholipid scramblase family, member 5 |
18895 |
0.21 |
| chr8_25465368_25465990 | 6.12 |
Gm10689 |
predicted gene 10689 |
11406 |
0.11 |
| chr1_107975730_107976183 | 5.99 |
Gm28282 |
predicted gene 28282 |
12649 |
0.2 |
| chr10_105689577_105689728 | 5.98 |
n-R5s80 |
nuclear encoded rRNA 5S 80 |
114545 |
0.05 |
| chr2_63669900_63670081 | 5.89 |
Gm23503 |
predicted gene, 23503 |
237796 |
0.02 |
| chr17_90639234_90639409 | 5.74 |
Nrxn1 |
neurexin I |
9097 |
0.28 |
| chr9_23580197_23580362 | 5.72 |
Gm3011 |
predicted gene 3011 |
196213 |
0.03 |
| chr4_63459173_63459841 | 5.71 |
Whrn |
whirlin |
1795 |
0.32 |
| chr16_72693663_72693924 | 5.68 |
Robo1 |
roundabout guidance receptor 1 |
30589 |
0.26 |
| chr3_52007190_52007930 | 5.63 |
Gm37465 |
predicted gene, 37465 |
3535 |
0.16 |
| chr5_3450585_3450801 | 5.62 |
Cdk6 |
cyclin-dependent kinase 6 |
20256 |
0.14 |
| chr1_41814359_41814918 | 5.61 |
Gm29260 |
predicted gene 29260 |
32830 |
0.25 |
| chr12_12254219_12254410 | 5.53 |
Fam49a |
family with sequence similarity 49, member A |
7825 |
0.29 |
| chr9_9583092_9583306 | 5.53 |
Gm46102 |
predicted gene, 46102 |
108 |
0.98 |
| chr7_49490873_49491264 | 5.51 |
Gm38059 |
predicted gene, 38059 |
21356 |
0.22 |
| chr13_83717468_83717619 | 5.49 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
3838 |
0.16 |
| chrX_14259705_14259856 | 5.49 |
Gm14867 |
predicted gene 14867 |
29959 |
0.19 |
| chr16_50585962_50586298 | 5.29 |
4930542D17Rik |
RIKEN cDNA 4930542D17 gene |
4373 |
0.22 |
| chr19_21775784_21775935 | 5.21 |
Cemip2 |
cell migration inducing hyaluronidase 2 |
2483 |
0.31 |
| chr7_72592750_72592946 | 5.15 |
Gm37620 |
predicted gene, 37620 |
9141 |
0.21 |
| chr1_53940244_53940474 | 5.15 |
Gm24251 |
predicted gene, 24251 |
53418 |
0.13 |
| chr4_45754240_45754391 | 5.13 |
Gm12409 |
predicted gene 12409 |
9908 |
0.15 |
| chr16_90353067_90353277 | 5.13 |
Gm49708 |
predicted gene, 49708 |
3530 |
0.17 |
| chr15_88290879_88291033 | 5.08 |
B230214G05Rik |
RIKEN cDNA B230214G05 gene |
23918 |
0.21 |
| chr1_132653907_132654070 | 4.99 |
Nfasc |
neurofascin |
16897 |
0.21 |
| chr18_33461781_33461964 | 4.93 |
Nrep |
neuronal regeneration related protein |
1563 |
0.38 |
| chr7_70104218_70104369 | 4.91 |
Gm35325 |
predicted gene, 35325 |
103142 |
0.06 |
| chr7_96595804_96596006 | 4.91 |
Gm15414 |
predicted gene 15414 |
33164 |
0.19 |
| chr2_22620080_22620281 | 4.91 |
Gad2 |
glutamic acid decarboxylase 2 |
2025 |
0.23 |
| chr8_62284797_62285008 | 4.89 |
Gm2961 |
predicted gene 2961 |
90690 |
0.09 |
| chr1_60860670_60860871 | 4.89 |
Gm38137 |
predicted gene, 38137 |
19568 |
0.12 |
| chr3_73046342_73046524 | 4.89 |
Gm37056 |
predicted gene, 37056 |
2594 |
0.26 |
| chr1_137606381_137606640 | 4.86 |
Gm37903 |
predicted gene, 37903 |
75280 |
0.1 |
| chr18_30509304_30509455 | 4.83 |
Gm7936 |
predicted pseudogene 7936 |
14006 |
0.19 |
| chr2_36010840_36011316 | 4.82 |
Ttll11 |
tubulin tyrosine ligase-like family, member 11 |
31165 |
0.12 |
| chr3_134302602_134302962 | 4.77 |
Gm43559 |
predicted gene 43559 |
30272 |
0.13 |
| chr18_10942003_10942165 | 4.74 |
Gm7575 |
predicted gene 7575 |
13200 |
0.22 |
| chr12_5250062_5250371 | 4.72 |
Gm48532 |
predicted gene, 48532 |
14252 |
0.23 |
| chr3_78908557_78908731 | 4.67 |
Gm10291 |
predicted pseudogene 10291 |
8614 |
0.18 |
| chr7_84285970_84286121 | 4.67 |
Arnt2 |
aryl hydrocarbon receptor nuclear translocator 2 |
3651 |
0.21 |
| chr1_25186041_25186192 | 4.62 |
Adgrb3 |
adhesion G protein-coupled receptor B3 |
18607 |
0.17 |
| chr17_45279348_45279645 | 4.52 |
Gm24979 |
predicted gene, 24979 |
69530 |
0.1 |
| chr10_97009843_97009994 | 4.51 |
4930556N09Rik |
RIKEN cDNA 4930556N09 gene |
25850 |
0.2 |
| chr10_102238597_102238748 | 4.46 |
Mgat4c |
MGAT4 family, member C |
79593 |
0.1 |
| chr11_15649048_15649299 | 4.46 |
Gm12010 |
predicted gene 12010 |
166867 |
0.04 |
| chr16_67579226_67579420 | 4.45 |
Gm15828 |
predicted gene 15828 |
13903 |
0.26 |
| chr3_41195114_41195652 | 4.44 |
Gm40038 |
predicted gene, 40038 |
22720 |
0.2 |
| chr10_79613505_79614566 | 4.43 |
C2cd4c |
C2 calcium-dependent domain containing 4C |
10 |
0.95 |
| chr18_69607136_69607287 | 4.41 |
Tcf4 |
transcription factor 4 |
5233 |
0.3 |
| chr19_37090377_37090698 | 4.37 |
Gm22714 |
predicted gene, 22714 |
58625 |
0.1 |
| chr6_47228750_47229021 | 4.36 |
Cntnap2 |
contactin associated protein-like 2 |
15502 |
0.28 |
| chr17_56510062_56510456 | 4.34 |
Znrf4 |
zinc and ring finger 4 |
2172 |
0.23 |
| chr6_111622889_111623040 | 4.25 |
Gm22093 |
predicted gene, 22093 |
19712 |
0.28 |
| chr18_25378079_25378424 | 4.20 |
Gm16558 |
predicted gene 16558 |
13550 |
0.27 |
| chr5_37065180_37065358 | 4.17 |
Jakmip1 |
janus kinase and microtubule interacting protein 1 |
14353 |
0.14 |
| chr14_11863272_11863596 | 4.16 |
Gm48601 |
predicted gene, 48601 |
4160 |
0.28 |
| chr2_51087621_51088075 | 4.14 |
Rnd3 |
Rho family GTPase 3 |
61246 |
0.13 |
| chr10_111099836_111100043 | 4.09 |
Gm48851 |
predicted gene, 48851 |
2932 |
0.22 |
| chr11_78786013_78786424 | 4.02 |
Gm23840 |
predicted gene, 23840 |
31696 |
0.11 |
| chr8_64346530_64346681 | 4.00 |
Gm35521 |
predicted gene, 35521 |
70495 |
0.12 |
| chr1_157888794_157888975 | 3.99 |
Gm38256 |
predicted gene, 38256 |
51501 |
0.17 |
| chr2_105719066_105719217 | 3.95 |
Gm13954 |
predicted gene 13954 |
10142 |
0.18 |
| chr7_49071017_49071168 | 3.93 |
Gm45207 |
predicted gene 45207 |
8511 |
0.19 |
| chr6_61043265_61043754 | 3.91 |
Gm43892 |
predicted gene, 43892 |
1484 |
0.4 |
| chr1_81593210_81593372 | 3.90 |
Gm6198 |
predicted gene 6198 |
35808 |
0.21 |
| chr16_66969715_66969926 | 3.86 |
Cadm2 |
cell adhesion molecule 2 |
16486 |
0.28 |
| chr13_83727309_83727854 | 3.83 |
C130071C03Rik |
RIKEN cDNA C130071C03 gene |
525 |
0.66 |
| chr2_84289370_84289521 | 3.81 |
Gm13711 |
predicted gene 13711 |
28866 |
0.19 |
| chr3_144378161_144378542 | 3.80 |
Gm5857 |
predicted gene 5857 |
49321 |
0.12 |
| chr14_78288901_78289283 | 3.79 |
Tnfsf11 |
tumor necrosis factor (ligand) superfamily, member 11 |
18951 |
0.17 |
| chr3_48429497_48429662 | 3.76 |
1700018B24Rik |
RIKEN cDNA 1700018B24 gene |
179523 |
0.03 |
| chr1_98192584_98192735 | 3.76 |
Gm29461 |
predicted gene 29461 |
19011 |
0.19 |
| chr3_16657689_16657840 | 3.75 |
Gm26485 |
predicted gene, 26485 |
165548 |
0.04 |
| chr13_110114248_110114399 | 3.70 |
Rab3c |
RAB3C, member RAS oncogene family |
165827 |
0.03 |
| chr14_89062657_89062861 | 3.63 |
Gm36946 |
predicted gene, 36946 |
288266 |
0.01 |
| chrX_61154998_61155204 | 3.59 |
Gm24396 |
predicted gene, 24396 |
9739 |
0.16 |
| chr4_72382144_72382349 | 3.55 |
Gm11235 |
predicted gene 11235 |
160420 |
0.04 |
| chr3_17797861_17798083 | 3.55 |
Mir124-2hg |
Mir124-2 host gene (non-protein coding) |
2228 |
0.25 |
| chr7_67607781_67607932 | 3.46 |
Lrrc28 |
leucine rich repeat containing 28 |
594 |
0.68 |
| chr4_22498680_22498974 | 3.44 |
Gm30731 |
predicted gene, 30731 |
8279 |
0.16 |
| chr2_68366435_68366739 | 3.41 |
Stk39 |
serine/threonine kinase 39 |
68 |
0.98 |
| chr8_48322765_48322916 | 3.40 |
Tenm3 |
teneurin transmembrane protein 3 |
22741 |
0.23 |
| chr5_135834810_135834968 | 3.39 |
Gm23268 |
predicted gene, 23268 |
2483 |
0.17 |
| chr15_4375139_4375310 | 3.38 |
Plcxd3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
267 |
0.95 |
| chr4_136820408_136820559 | 3.38 |
Ephb2 |
Eph receptor B2 |
15360 |
0.19 |
| chr10_106609605_106610022 | 3.34 |
4930532I03Rik |
RIKEN cDNA 4930532I03 gene |
7333 |
0.29 |
| chr5_15993645_15993826 | 3.32 |
Gm43000 |
predicted gene 43000 |
4104 |
0.23 |
| chr3_149088825_149089063 | 3.31 |
Gm25127 |
predicted gene, 25127 |
60012 |
0.12 |
| chr15_65479129_65479280 | 3.31 |
Gm49243 |
predicted gene, 49243 |
208944 |
0.02 |
| chr12_112238743_112239296 | 3.29 |
Gm20368 |
predicted gene, 20368 |
33866 |
0.14 |
| chr12_50122622_50122808 | 3.29 |
Gm40418 |
predicted gene, 40418 |
2406 |
0.45 |
| chr6_51849740_51849921 | 3.25 |
Skap2 |
src family associated phosphoprotein 2 |
22099 |
0.22 |
| chr17_37078905_37079056 | 3.25 |
Olfr90 |
olfactory receptor 90 |
7256 |
0.08 |
| chr5_71604258_71604409 | 3.20 |
Gabra4 |
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
53487 |
0.13 |
| chr7_55332148_55332299 | 3.19 |
Gm44898 |
predicted gene 44898 |
56287 |
0.13 |
| chr12_58670274_58670654 | 3.19 |
Gm18873 |
predicted gene, 18873 |
93442 |
0.08 |
| chr6_141485321_141485684 | 3.18 |
Slco1c1 |
solute carrier organic anion transporter family, member 1c1 |
38866 |
0.17 |
| chr19_42904302_42904453 | 3.18 |
Hps1 |
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
124399 |
0.05 |
| chr5_119080735_119081067 | 3.17 |
1700081H04Rik |
RIKEN cDNA 1700081H04 gene |
27333 |
0.21 |
| chr1_137453589_137453740 | 3.17 |
Gm37903 |
predicted gene, 37903 |
77566 |
0.09 |
| chr2_20072451_20072602 | 3.16 |
Gm13360 |
predicted gene 13360 |
34632 |
0.18 |
| chr1_42231020_42231171 | 3.14 |
Gm9915 |
predicted gene 9915 |
1368 |
0.42 |
| chr6_94751135_94751286 | 3.14 |
Gm43997 |
predicted gene, 43997 |
48222 |
0.11 |
| chr10_18458247_18458479 | 3.13 |
Nhsl1 |
NHS-like 1 |
11525 |
0.24 |
| chrX_52369390_52369575 | 3.08 |
Mir6384 |
microRNA 6384 |
8699 |
0.24 |
| chr5_21733359_21733517 | 3.08 |
Pmpcb |
peptidase (mitochondrial processing) beta |
3703 |
0.16 |
| chr19_37991765_37991916 | 3.08 |
Myof |
myoferlin |
24714 |
0.17 |
| chr3_102353639_102353854 | 3.08 |
Gm43242 |
predicted gene 43242 |
15416 |
0.18 |
| chr10_95790795_95791109 | 3.04 |
4732465J04Rik |
RIKEN cDNA 4732465J04 gene |
12035 |
0.12 |
| chr3_105481595_105481754 | 3.03 |
Gm43847 |
predicted gene 43847 |
21233 |
0.17 |
| chr15_52565416_52565897 | 3.01 |
Gm23267 |
predicted gene, 23267 |
66000 |
0.11 |
| chr7_96505702_96505853 | 3.01 |
Tenm4 |
teneurin transmembrane protein 4 |
16621 |
0.24 |
| chr18_25016662_25016880 | 3.00 |
Fhod3 |
formin homology 2 domain containing 3 |
107590 |
0.07 |
| chr10_31051366_31051517 | 2.99 |
Gm30676 |
predicted gene, 30676 |
14793 |
0.23 |
| chr2_106499568_106499880 | 2.98 |
Gm14015 |
predicted gene 14015 |
23379 |
0.23 |
| chr3_34967971_34968243 | 2.97 |
Mir6378 |
microRNA 6378 |
45456 |
0.18 |
| chr5_13312458_13312623 | 2.95 |
Sema3a |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
84244 |
0.09 |
| chr15_88978956_88979127 | 2.94 |
Mlc1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human) |
34 |
0.95 |
| chr19_21549066_21549217 | 2.91 |
Gm3443 |
predicted gene 3443 |
3063 |
0.3 |
| chr11_57070540_57070691 | 2.91 |
Gria1 |
glutamate receptor, ionotropic, AMPA1 (alpha 1) |
57569 |
0.17 |
| chr7_118593604_118593757 | 2.89 |
B230311B06Rik |
RIKEN cDNA B230311B06 gene |
1164 |
0.39 |
| chr13_90643930_90644265 | 2.87 |
Gm36966 |
predicted gene, 36966 |
10012 |
0.25 |
| chr18_73867474_73867843 | 2.85 |
Mro |
maestro |
3986 |
0.27 |
| chr2_123507243_123507394 | 2.85 |
Gm13988 |
predicted gene 13988 |
233394 |
0.02 |
| chr6_34346643_34346822 | 2.84 |
Akr1b8 |
aldo-keto reductase family 1, member B8 |
7387 |
0.14 |
| chr1_12693326_12693565 | 2.83 |
Sulf1 |
sulfatase 1 |
913 |
0.57 |
| chr2_141312213_141312551 | 2.83 |
Macrod2os1 |
mono-ADP ribosylhydrolase 2, opposite strand 1 |
25391 |
0.25 |
| chr10_96980838_96981021 | 2.82 |
Gm33981 |
predicted gene, 33981 |
22058 |
0.21 |
| chr1_172242412_172242774 | 2.79 |
Gm37950 |
predicted gene, 37950 |
5302 |
0.1 |
| chr7_46162072_46162347 | 2.78 |
Abcc8 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
17777 |
0.11 |
| chr14_59819467_59819618 | 2.74 |
Gm19716 |
predicted gene, 19716 |
176994 |
0.03 |
| chr18_81251894_81252538 | 2.69 |
Gm30192 |
predicted gene, 30192 |
12475 |
0.22 |
| chr8_125669580_125669961 | 2.69 |
Map10 |
microtubule-associated protein 10 |
48 |
0.98 |
| chr2_107734427_107734615 | 2.68 |
Gm9864 |
predicted gene 9864 |
133982 |
0.05 |
| chr1_92680883_92681479 | 2.65 |
Otos |
otospiralin |
32340 |
0.09 |
| chr1_153660852_153661003 | 2.63 |
Rgs8 |
regulator of G-protein signaling 8 |
154 |
0.94 |
| chr15_51104377_51104528 | 2.63 |
Gm48913 |
predicted gene, 48913 |
206813 |
0.02 |
| chr13_29015675_29016011 | 2.62 |
A330102I10Rik |
RIKEN cDNA A330102I10 gene |
450 |
0.87 |
| chr1_43299581_43299777 | 2.62 |
Gm29040 |
predicted gene 29040 |
3246 |
0.26 |
| chr15_102510366_102511154 | 2.61 |
Map3k12 |
mitogen-activated protein kinase kinase kinase 12 |
32 |
0.95 |
| chr13_97395330_97395481 | 2.59 |
Gm33447 |
predicted gene, 33447 |
19171 |
0.17 |
| chr8_102538158_102538348 | 2.57 |
Gm45422 |
predicted gene 45422 |
3608 |
0.28 |
| chr2_137092508_137093266 | 2.57 |
Jag1 |
jagged 1 |
6938 |
0.29 |
| chr14_60377670_60378016 | 2.56 |
Amer2 |
APC membrane recruitment 2 |
151 |
0.96 |
| chr16_45690308_45690465 | 2.56 |
Tmprss7 |
transmembrane serine protease 7 |
771 |
0.57 |
| chr1_194613964_194614514 | 2.53 |
Plxna2 |
plexin A2 |
3979 |
0.24 |
| chr18_12740759_12741006 | 2.53 |
Cabyr |
calcium-binding tyrosine-(Y)-phosphorylation regulated (fibrousheathin 2) |
429 |
0.75 |
| chr8_95703143_95704225 | 2.52 |
Ndrg4 |
N-myc downstream regulated gene 4 |
614 |
0.57 |
| chr3_123447332_123448076 | 2.48 |
Gm35065 |
predicted gene, 35065 |
257 |
0.79 |
| chr16_63753151_63753409 | 2.46 |
Gm22769 |
predicted gene, 22769 |
5746 |
0.33 |
| chr4_14106224_14106501 | 2.46 |
Gm24068 |
predicted gene, 24068 |
26422 |
0.21 |
| chr4_148360888_148361043 | 2.43 |
Gm13200 |
predicted gene 13200 |
1505 |
0.37 |
| chr2_91480915_91481066 | 2.43 |
Lrp4 |
low density lipoprotein receptor-related protein 4 |
4671 |
0.18 |
| chr14_100283490_100283858 | 2.43 |
Klf12 |
Kruppel-like factor 12 |
810 |
0.61 |
| chr17_14164003_14164270 | 2.43 |
Gm49903 |
predicted gene, 49903 |
37839 |
0.12 |
| chr3_56955404_56955772 | 2.43 |
Gm22269 |
predicted gene, 22269 |
98562 |
0.08 |
| chr1_94503721_94503899 | 2.41 |
Gm7895 |
predicted gene 7895 |
33923 |
0.23 |
| chr15_52776567_52776718 | 2.41 |
Gm34794 |
predicted gene, 34794 |
9625 |
0.28 |
| chr13_107541201_107541459 | 2.41 |
Gm32004 |
predicted gene, 32004 |
23993 |
0.2 |
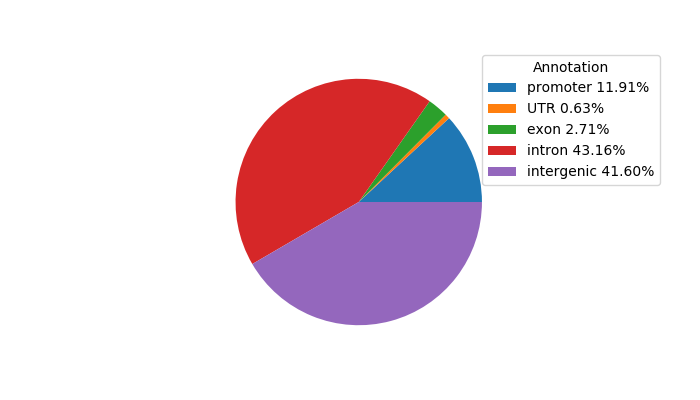
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0061346 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 1.1 | 3.3 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.8 | 2.4 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.8 | 7.1 | GO:0086036 | regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.8 | 2.3 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.7 | 2.0 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.6 | 2.6 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.6 | 1.7 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.5 | 2.0 | GO:1904398 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) positive regulation of neuromuscular junction development(GO:1904398) |
| 0.5 | 1.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.5 | 2.8 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.4 | 1.8 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.4 | 1.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.4 | 1.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.4 | 2.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.4 | 2.7 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.3 | 2.0 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 1.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 | 2.6 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.3 | 1.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.3 | 0.6 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.3 | 0.9 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) |
| 0.3 | 1.8 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 1.2 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.3 | 1.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.3 | 1.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.3 | 0.8 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.2 | 1.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.2 | 0.7 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.2 | 0.7 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.2 | 1.1 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.2 | 0.4 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.2 | 0.6 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.2 | 2.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 0.6 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.2 | 0.7 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 0.9 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 0.7 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.2 | 1.4 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.2 | 1.4 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.2 | 0.8 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.2 | 0.8 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.2 | 0.8 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 0.2 | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.2 | 0.5 | GO:0015755 | fructose transport(GO:0015755) |
| 0.2 | 1.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.2 | 0.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 1.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.9 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 0.7 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.7 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 0.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.7 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.1 | 0.5 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 0.4 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.5 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.1 | 0.3 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 1.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.4 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.3 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.3 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.3 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 | 0.5 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.1 | 0.9 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.9 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.3 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.3 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 6.5 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 0.3 | GO:1903998 | regulation of eating behavior(GO:1903998) negative regulation of eating behavior(GO:1903999) |
| 0.1 | 0.4 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 | 0.4 | GO:0060509 | Type I pneumocyte differentiation(GO:0060509) |
| 0.1 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.3 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 0.3 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.5 | GO:0099625 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 | 0.1 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.7 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.7 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 0.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.6 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.5 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 0.2 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.3 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.4 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.5 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 2.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.5 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.6 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.1 | 0.1 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.1 | 0.1 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.2 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 | 0.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.2 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.1 | GO:1903802 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.1 | 1.0 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.1 | 0.5 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.1 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.2 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.1 | 1.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.1 | 0.2 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:1901844 | cell communication by electrical coupling involved in cardiac conduction(GO:0086064) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.0 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:2000173 | negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 1.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.7 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.3 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.0 | 0.2 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.0 | 1.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.5 | GO:0014741 | negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 | 0.0 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) nucleotide transmembrane transport(GO:1901679) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.8 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.2 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.5 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.0 | 0.0 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.1 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.0 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.0 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.0 | GO:0021825 | substrate-dependent cerebral cortex tangential migration(GO:0021825) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:0036376 | sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0003309 | type B pancreatic cell differentiation(GO:0003309) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.0 | 0.6 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0072051 | juxtaglomerular apparatus development(GO:0072051) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.0 | 0.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.0 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.1 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.0 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0044091 | membrane biogenesis(GO:0044091) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.0 | GO:0036296 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) cellular response to hyperoxia(GO:0071455) |
| 0.0 | 0.0 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0036491 | regulation of translation in response to endoplasmic reticulum stress(GO:0036490) regulation of translation initiation in response to endoplasmic reticulum stress(GO:0036491) eiF2alpha phosphorylation in response to endoplasmic reticulum stress(GO:0036492) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:0045914 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.0 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.0 | GO:0002905 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.5 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.0 | 0.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.0 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.3 | GO:0002438 | acute inflammatory response to antigenic stimulus(GO:0002438) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.0 | 0.1 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.0 | 0.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) |
| 0.0 | 0.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0071688 | skeletal muscle myosin thick filament assembly(GO:0030241) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0072366 | regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0051709 | regulation of killing of cells of other organism(GO:0051709) |
| 0.0 | 0.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.0 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.0 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.0 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.0 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.0 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.0 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.0 | 0.0 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0072177 | mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.0 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.0 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.4 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.0 | 0.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.0 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.0 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.0 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.0 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.8 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.0 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:0010535 | positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.0 | 0.0 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.0 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.0 | 0.1 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.1 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.0 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.0 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.6 | 2.4 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.5 | 1.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.5 | 1.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.4 | 1.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.4 | 2.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.3 | 6.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.3 | 1.6 | GO:0071547 | piP-body(GO:0071547) |
| 0.3 | 2.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 0.7 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.2 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 3.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 4.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 0.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 1.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 1.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 4.9 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.8 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 1.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 1.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.0 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 1.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 2.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.0 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.7 | 3.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.7 | 6.2 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.6 | 2.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 1.6 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 1.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 1.6 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.5 | 1.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 1.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 2.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 0.9 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.3 | 1.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 0.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.2 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 0.9 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.2 | 1.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 1.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 1.5 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.2 | 1.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 2.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.5 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 2.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.1 | GO:0016662 | oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.3 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 2.3 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 0.2 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 0.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.2 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0001031 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 2.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 1.7 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0008412 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 2.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.4 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.4 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:1901567 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.0 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0052717 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.6 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 1.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.0 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.9 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.1 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.0 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.0 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 3.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 1.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 5.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 0.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 2.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.0 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 0.8 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.7 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.9 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.0 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |