Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
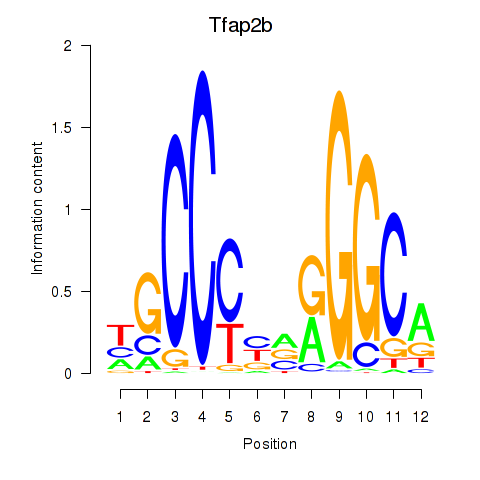
Results for Tfap2b
Z-value: 1.26
Transcription factors associated with Tfap2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2b
|
ENSMUSG00000025927.7 | transcription factor AP-2 beta |
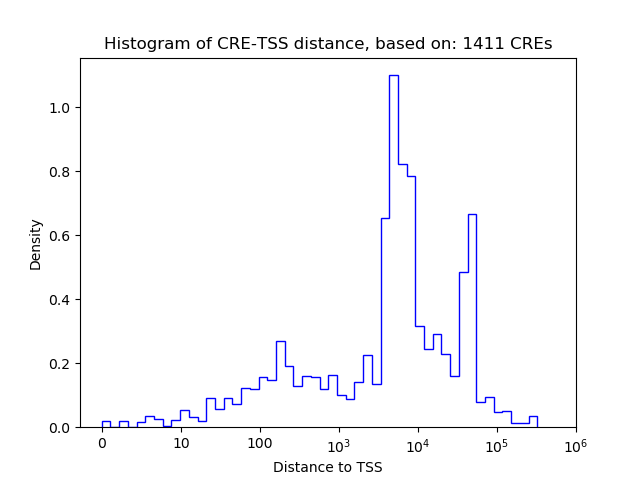
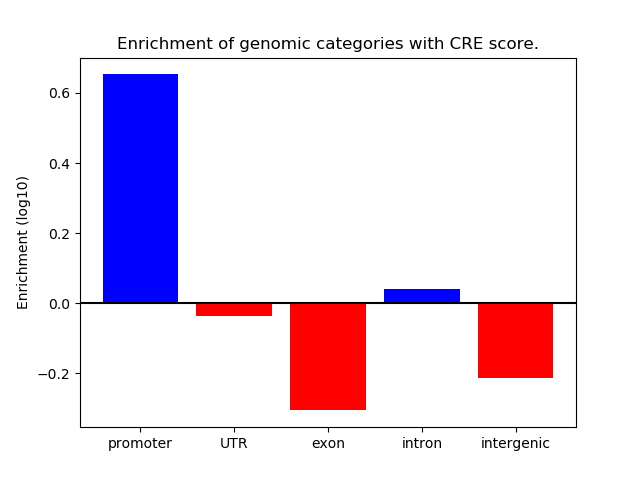
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_19226762_19226913 | Tfap2b | 12958 | 0.186524 | 0.10 | 4.6e-01 | Click! |
| chr1_19217099_19217250 | Tfap2b | 3295 | 0.253303 | -0.04 | 7.7e-01 | Click! |
| chr1_19209073_19209255 | Tfap2b | 204 | 0.944914 | -0.03 | 8.3e-01 | Click! |
| chr1_19217371_19217862 | Tfap2b | 3737 | 0.240551 | -0.02 | 8.8e-01 | Click! |
| chr1_19205838_19205989 | Tfap2b | 3001 | 0.265345 | -0.01 | 9.3e-01 | Click! |
Activity of the Tfap2b motif across conditions
Conditions sorted by the z-value of the Tfap2b motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
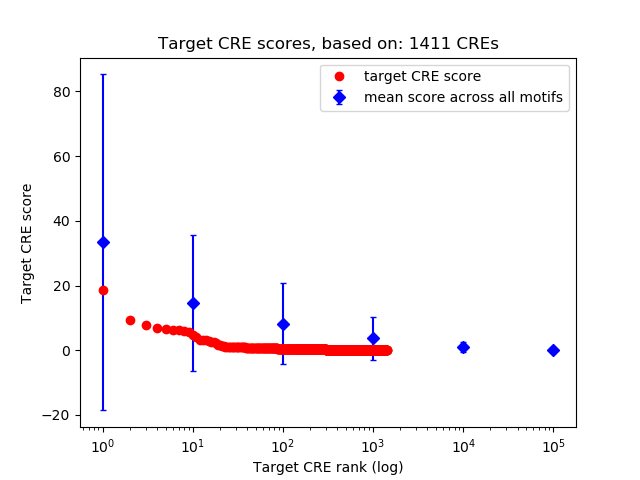
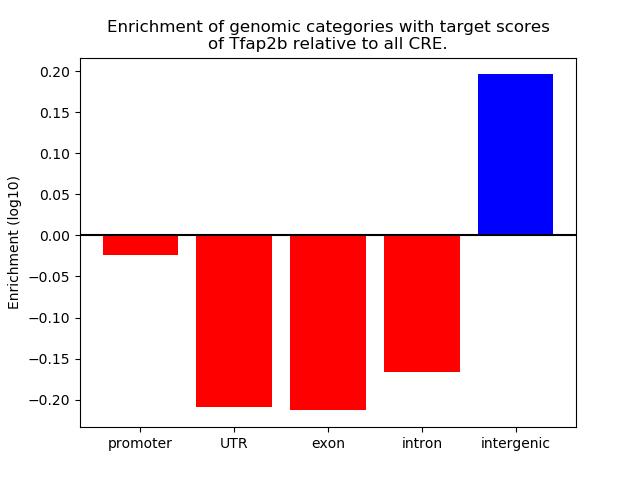
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_3117365_3118622 | 18.50 |
Vmn1r238 |
vomeronasal 1 receptor, 238 |
5419 |
0.17 |
| chr18_3116815_3117174 | 9.28 |
Vmn1r238 |
vomeronasal 1 receptor, 238 |
6418 |
0.16 |
| chr18_20621651_20622570 | 7.89 |
Gm16090 |
predicted gene 16090 |
43150 |
0.11 |
| chr7_12010381_12010846 | 6.83 |
Vmn1r-ps60 |
vomeronasal 1 receptor, pseudogene 60 |
5547 |
0.07 |
| chr18_3116148_3116568 | 6.68 |
Vmn1r238 |
vomeronasal 1 receptor, 238 |
7054 |
0.16 |
| chr18_20616739_20617639 | 6.31 |
Gm16090 |
predicted gene 16090 |
48071 |
0.1 |
| chr18_3118971_3119257 | 6.29 |
Vmn1r238 |
vomeronasal 1 receptor, 238 |
4298 |
0.18 |
| chr7_12012707_12013031 | 5.88 |
Vmn1r-ps60 |
vomeronasal 1 receptor, pseudogene 60 |
7803 |
0.06 |
| chr18_3119520_3119704 | 5.57 |
Vmn1r238 |
vomeronasal 1 receptor, 238 |
3800 |
0.19 |
| chr7_12012353_12012504 | 4.80 |
Vmn1r-ps60 |
vomeronasal 1 receptor, pseudogene 60 |
7362 |
0.06 |
| chr18_20625060_20625736 | 4.03 |
Gm16090 |
predicted gene 16090 |
39862 |
0.12 |
| chr18_3115836_3115987 | 3.31 |
Vmn1r238 |
vomeronasal 1 receptor, 238 |
7501 |
0.16 |
| chr4_3165180_3165331 | 3.26 |
Vmn1r2 |
vomeronasal 1 receptor 2 |
2065 |
0.29 |
| chr7_12011626_12011813 | 3.18 |
Vmn1r-ps60 |
vomeronasal 1 receptor, pseudogene 60 |
6653 |
0.07 |
| chr18_3115468_3115789 | 2.86 |
Vmn1r238 |
vomeronasal 1 receptor, 238 |
7784 |
0.16 |
| chr18_20615543_20616002 | 2.47 |
Gm16090 |
predicted gene 16090 |
49488 |
0.1 |
| chr18_3118775_3118949 | 2.44 |
Vmn1r238 |
vomeronasal 1 receptor, 238 |
4550 |
0.18 |
| chr4_3176842_3177034 | 2.30 |
Vmn1r2 |
vomeronasal 1 receptor 2 |
4855 |
0.19 |
| chr18_80287426_80288075 | 1.50 |
Gm37015 |
predicted gene, 37015 |
15930 |
0.11 |
| chr13_72833435_72833586 | 1.47 |
D730050B12Rik |
RIKEN cDNA D730050B12 gene |
16747 |
0.24 |
| chr18_20627927_20628180 | 1.37 |
Gm16090 |
predicted gene 16090 |
37207 |
0.12 |
| chr5_15672583_15672734 | 1.22 |
Speer4cos |
spermatogenesis associated glutamate (E)-rich protein 4C, opposite strand transcript |
8052 |
0.16 |
| chr7_102025253_102025404 | 1.06 |
Rnf121 |
ring finger protein 121 |
859 |
0.42 |
| chr18_20629240_20629391 | 1.05 |
Gm16090 |
predicted gene 16090 |
35945 |
0.13 |
| chr5_15043056_15043231 | 1.04 |
Gm17019 |
predicted gene 17019 |
10145 |
0.25 |
| chr7_74687595_74687746 | 1.01 |
Gm7726 |
predicted gene 7726 |
12186 |
0.26 |
| chr5_14949256_14949587 | 1.00 |
Speer4e |
spermatogenesis associated glutamate (E)-rich protein 4e |
10992 |
0.19 |
| chr3_53984780_53984955 | 0.98 |
Gm8109 |
predicted gene 8109 |
36297 |
0.15 |
| chr18_20624320_20624587 | 0.96 |
Gm16090 |
predicted gene 16090 |
40807 |
0.12 |
| chr5_15551399_15551801 | 0.94 |
Gm21190 |
predicted gene, 21190 |
22378 |
0.13 |
| chr6_120539180_120540180 | 0.93 |
Hdhd5 |
haloacid dehalogenase like hydrolase domain containing 5 |
8361 |
0.12 |
| chr18_20620820_20620971 | 0.91 |
Gm16090 |
predicted gene 16090 |
44365 |
0.11 |
| chr4_136865069_136865433 | 0.90 |
C1qb |
complement component 1, q subcomponent, beta polypeptide |
20936 |
0.14 |
| chr10_60692133_60692313 | 0.87 |
Cdh23 |
cadherin 23 (otocadherin) |
4267 |
0.26 |
| chr1_57218512_57218908 | 0.86 |
BC055402 |
cDNA sequence BC055402 |
3717 |
0.29 |
| chr5_15608945_15609307 | 0.86 |
Gm30613 |
predicted gene, 30613 |
4060 |
0.18 |
| chr17_61302066_61302300 | 0.86 |
Obox6-ps1 |
oocyte specific homeobox 6, pseudogene 1 |
58390 |
0.15 |
| chr18_46484000_46484192 | 0.84 |
Gm24076 |
predicted gene, 24076 |
10072 |
0.13 |
| chr18_20623022_20623191 | 0.82 |
Gm16090 |
predicted gene 16090 |
42154 |
0.12 |
| chr13_54693536_54694669 | 0.81 |
Rnf44 |
ring finger protein 44 |
195 |
0.91 |
| chr2_109693106_109694421 | 0.81 |
Bdnf |
brain derived neurotrophic factor |
200 |
0.94 |
| chr1_194354828_194354979 | 0.81 |
4930503O07Rik |
RIKEN cDNA 4930503O07 gene |
132144 |
0.05 |
| chr18_20626775_20626931 | 0.80 |
Gm16090 |
predicted gene 16090 |
38407 |
0.12 |
| chr18_20626425_20626742 | 0.79 |
Gm16090 |
predicted gene 16090 |
38677 |
0.12 |
| chr5_14988977_14989543 | 0.78 |
Gm10354 |
predicted gene 10354 |
10325 |
0.21 |
| chr6_107713625_107713778 | 0.77 |
4933431M02Rik |
RIKEN cDNA 4933431M02 gene |
85927 |
0.09 |
| chr6_75281622_75281787 | 0.76 |
Gm6210 |
predicted gene 6210 |
16355 |
0.25 |
| chr18_20622786_20622961 | 0.73 |
Gm16090 |
predicted gene 16090 |
42387 |
0.11 |
| chr9_10269175_10269351 | 0.73 |
Gm24496 |
predicted gene, 24496 |
21324 |
0.22 |
| chr17_79605921_79606072 | 0.72 |
Rmdn2 |
regulator of microtubule dynamics 2 |
5867 |
0.28 |
| chr2_156666341_156666675 | 0.71 |
Gm14172 |
predicted gene 14172 |
33040 |
0.1 |
| chr2_74682809_74683583 | 0.71 |
Gm28309 |
predicted gene 28309 |
250 |
0.69 |
| chr1_21960934_21962001 | 0.70 |
Kcnq5 |
potassium voltage-gated channel, subfamily Q, member 5 |
114 |
0.98 |
| chr1_138837789_138838010 | 0.70 |
Lhx9 |
LIM homeobox protein 9 |
4240 |
0.17 |
| chr2_59562684_59563070 | 0.69 |
Gm13552 |
predicted gene 13552 |
1807 |
0.42 |
| chr7_67372280_67373557 | 0.68 |
Mef2a |
myocyte enhancer factor 2A |
60 |
0.97 |
| chr18_20618066_20618220 | 0.67 |
Gm16090 |
predicted gene 16090 |
47117 |
0.11 |
| chr19_43616167_43616368 | 0.66 |
Nkx2-3 |
NK2 homeobox 3 |
3942 |
0.13 |
| chr2_126555283_126555434 | 0.65 |
Slc27a2 |
solute carrier family 27 (fatty acid transporter), member 2 |
770 |
0.66 |
| chr9_8900765_8902148 | 0.64 |
Pgr |
progesterone receptor |
988 |
0.68 |
| chr17_25609260_25609765 | 0.63 |
Lmf1 |
lipase maturation factor 1 |
30192 |
0.07 |
| chr14_122474708_122475243 | 0.63 |
2610035F20Rik |
RIKEN cDNA 2610035F20 gene |
224 |
0.69 |
| chr15_27814807_27814975 | 0.62 |
Trio |
triple functional domain (PTPRF interacting) |
8892 |
0.22 |
| chr2_32317120_32318698 | 0.62 |
Gm23363 |
predicted gene, 23363 |
356 |
0.45 |
| chr2_106692893_106694599 | 0.61 |
Mpped2 |
metallophosphoesterase domain containing 2 |
477 |
0.85 |
| chr7_68311669_68311820 | 0.61 |
4930405G09Rik |
RIKEN cDNA 4930405G09 gene |
5288 |
0.14 |
| chr18_20620535_20620686 | 0.61 |
Gm16090 |
predicted gene 16090 |
44650 |
0.11 |
| chr1_82634564_82634787 | 0.60 |
n-R5s213 |
nuclear encoded rRNA 5S 213 |
338 |
0.87 |
| chr18_20626992_20627160 | 0.60 |
Gm16090 |
predicted gene 16090 |
38184 |
0.12 |
| chr7_49560224_49560375 | 0.59 |
Nav2 |
neuron navigator 2 |
6407 |
0.28 |
| chr10_121475538_121476618 | 0.58 |
Rassf3 |
Ras association (RalGDS/AF-6) domain family member 3 |
269 |
0.86 |
| chr17_35425252_35425979 | 0.57 |
H2-Q6 |
histocompatibility 2, Q region locus 6 |
738 |
0.41 |
| chr4_147349805_147350010 | 0.57 |
Zfp978 |
zinc finger protein 978 |
11396 |
0.13 |
| chr10_126978694_126979699 | 0.57 |
Ctdsp2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
167 |
0.89 |
| chr5_122092798_122093169 | 0.57 |
Myl2 |
myosin, light polypeptide 2, regulatory, cardiac, slow |
7968 |
0.14 |
| chr17_47878037_47879117 | 0.56 |
Foxp4 |
forkhead box P4 |
376 |
0.81 |
| chr3_132949340_132950694 | 0.56 |
Npnt |
nephronectin |
26 |
0.97 |
| chr8_66486037_66486995 | 0.56 |
Tma16 |
translation machinery associated 16 |
3 |
0.97 |
| chr12_112125881_112126258 | 0.55 |
Mir203 |
microRNA 203 |
4811 |
0.13 |
| chr10_96460717_96460875 | 0.55 |
Gm8601 |
predicted gene 8601 |
15683 |
0.19 |
| chr3_122729485_122730960 | 0.55 |
Pde5a |
phosphodiesterase 5A, cGMP-specific |
0 |
0.98 |
| chr8_97633073_97633363 | 0.54 |
Gm7191 |
predicted gene 7191 |
66626 |
0.14 |
| chr2_44920846_44920997 | 0.54 |
Gtdc1 |
glycosyltransferase-like domain containing 1 |
6228 |
0.29 |
| chr8_122610722_122612153 | 0.54 |
Galns |
galactosamine (N-acetyl)-6-sulfate sulfatase |
26 |
0.7 |
| chr8_15027100_15028701 | 0.53 |
Gm37844 |
predicted gene, 37844 |
522 |
0.44 |
| chr9_23487032_23487234 | 0.53 |
Bmper |
BMP-binding endothelial regulator |
113201 |
0.07 |
| chr18_20627726_20627877 | 0.53 |
Gm16090 |
predicted gene 16090 |
37459 |
0.12 |
| chr9_91378183_91378872 | 0.53 |
Zic4 |
zinc finger protein of the cerebellum 4 |
115 |
0.94 |
| chr17_28553454_28553746 | 0.53 |
Clpsl2 |
colipase-like 2 |
4189 |
0.09 |
| chr3_130839108_130839269 | 0.53 |
Gm5982 |
predicted gene 5982 |
7785 |
0.12 |
| chr6_125157136_125157596 | 0.52 |
Iffo1 |
intermediate filament family orphan 1 |
4018 |
0.08 |
| chr9_59539422_59540161 | 0.51 |
Tmem202 |
transmembrane protein 202 |
56 |
0.75 |
| chr10_107271547_107272835 | 0.50 |
Lin7a |
lin-7 homolog A (C. elegans) |
203 |
0.96 |
| chr11_21997009_21997356 | 0.50 |
Otx1 |
orthodenticle homeobox 1 |
4433 |
0.28 |
| chr19_26977887_26978038 | 0.49 |
Gm50121 |
predicted gene, 50121 |
48019 |
0.16 |
| chr12_29871996_29872873 | 0.49 |
Myt1l |
myelin transcription factor 1-like |
20886 |
0.24 |
| chr5_125389083_125389845 | 0.49 |
Gm10382 |
predicted gene 10382 |
142 |
0.67 |
| chr2_179292721_179292959 | 0.49 |
Gm14293 |
predicted gene 14293 |
52350 |
0.14 |
| chr17_85628600_85628757 | 0.49 |
Six3 |
sine oculis-related homeobox 3 |
7647 |
0.15 |
| chr5_43032065_43032228 | 0.48 |
Gm43700 |
predicted gene 43700 |
6090 |
0.19 |
| chr15_76133386_76133744 | 0.47 |
BC024139 |
cDNA sequence BC024139 |
6969 |
0.09 |
| chr15_102257553_102258012 | 0.47 |
Rarg |
retinoic acid receptor, gamma |
265 |
0.82 |
| chr8_46740254_46741749 | 0.47 |
Irf2 |
interferon regulatory factor 2 |
735 |
0.53 |
| chr1_189343290_189344052 | 0.47 |
Kcnk2 |
potassium channel, subfamily K, member 2 |
36 |
0.76 |
| chr6_114968557_114970098 | 0.47 |
Vgll4 |
vestigial like family member 4 |
147 |
0.97 |
| chr6_52245954_52246438 | 0.46 |
Hoxa11os |
homeobox A11, opposite strand |
25 |
0.82 |
| chr16_90533266_90533417 | 0.46 |
Gm36001 |
predicted gene, 36001 |
17268 |
0.15 |
| chr12_108906675_108906826 | 0.46 |
Wdr25 |
WD repeat domain 25 |
5982 |
0.13 |
| chr16_17497025_17497176 | 0.46 |
Aifm3 |
apoptosis-inducing factor, mitochondrion-associated 3 |
2599 |
0.15 |
| chr13_110230204_110230355 | 0.45 |
Rab3c |
RAB3C, member RAS oncogene family |
49871 |
0.16 |
| chr7_57508956_57510254 | 0.45 |
Gabra5 |
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5 |
234 |
0.95 |
| chr5_36100815_36100966 | 0.45 |
Psapl1 |
prosaposin-like 1 |
103131 |
0.07 |
| chr10_107437084_107437235 | 0.45 |
Gm38117 |
predicted gene, 38117 |
200 |
0.95 |
| chr17_17827653_17828872 | 0.45 |
Spaca6 |
sperm acrosome associated 6 |
938 |
0.31 |
| chr5_123027620_123027774 | 0.44 |
Gm43412 |
predicted gene 43412 |
8303 |
0.09 |
| chr14_14345884_14346978 | 0.44 |
Il3ra |
interleukin 3 receptor, alpha chain |
1 |
0.93 |
| chr13_105256983_105257274 | 0.44 |
Rnf180 |
ring finger protein 180 |
13911 |
0.24 |
| chr11_5835203_5835354 | 0.44 |
Polm |
polymerase (DNA directed), mu |
1207 |
0.32 |
| chr4_10883845_10884023 | 0.43 |
2610301B20Rik |
RIKEN cDNA 2610301B20 gene |
4395 |
0.16 |
| chr16_73096919_73097070 | 0.43 |
4930500H12Rik |
RIKEN cDNA 4930500H12 gene |
1913 |
0.49 |
| chr13_41705506_41705657 | 0.43 |
Gm5082 |
predicted gene 5082 |
49884 |
0.12 |
| chr1_63273306_63273901 | 0.43 |
Gm24513 |
predicted gene, 24513 |
240 |
0.49 |
| chr4_120170416_120170874 | 0.43 |
Edn2 |
endothelin 2 |
9439 |
0.24 |
| chr1_18463225_18463376 | 0.43 |
Defb41 |
defensin beta 41 |
198162 |
0.02 |
| chr18_68711266_68711468 | 0.42 |
Mir6356 |
microRNA 6356 |
28033 |
0.21 |
| chr16_74182688_74182857 | 0.42 |
Gm49659 |
predicted gene, 49659 |
3938 |
0.24 |
| chr10_122985267_122986641 | 0.42 |
D630033A02Rik |
RIKEN cDNA D630033A02 gene |
123 |
0.65 |
| chr5_14914343_14915097 | 0.42 |
Gm9758 |
predicted gene 9758 |
133 |
0.97 |
| chr5_136711274_136711425 | 0.42 |
Myl10 |
myosin, light chain 10, regulatory |
13519 |
0.2 |
| chr19_42764540_42764691 | 0.41 |
Hps1 |
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
739 |
0.61 |
| chr9_22767838_22767989 | 0.41 |
Gm27639 |
predicted gene, 27639 |
4922 |
0.27 |
| chr6_87898144_87898295 | 0.41 |
Copg1 |
coatomer protein complex, subunit gamma 1 |
416 |
0.7 |
| chr3_40185875_40186026 | 0.41 |
1700017G19Rik |
RIKEN cDNA 1700017G19 gene |
323824 |
0.01 |
| chr5_90988282_90988433 | 0.40 |
Epgn |
epithelial mitogen |
39107 |
0.11 |
| chr4_154709923_154710074 | 0.40 |
Actrt2 |
actin-related protein T2 |
42131 |
0.11 |
| chr14_36646374_36646525 | 0.40 |
Gm17940 |
predicted gene, 17940 |
27171 |
0.2 |
| chr6_55335921_55336550 | 0.39 |
Aqp1 |
aquaporin 1 |
197 |
0.93 |
| chr2_32390923_32391074 | 0.39 |
Lcn2 |
lipocalin 2 |
2746 |
0.12 |
| chr2_31508651_31508802 | 0.39 |
Ass1 |
argininosuccinate synthetase 1 |
8000 |
0.18 |
| chr10_87586828_87586979 | 0.38 |
Pah |
phenylalanine hydroxylase |
40230 |
0.16 |
| chr5_15577027_15578025 | 0.38 |
Gm21083 |
predicted gene, 21083 |
193 |
0.93 |
| chr13_56769978_56770289 | 0.38 |
Gm45623 |
predicted gene 45623 |
16306 |
0.21 |
| chr3_116969934_116970085 | 0.38 |
Palmd |
palmdelphin |
1022 |
0.4 |
| chr16_40652871_40653153 | 0.37 |
Gm27887 |
predicted gene, 27887 |
285075 |
0.01 |
| chr19_3392510_3392690 | 0.37 |
Tesmin |
testis expressed metallothionein like |
3221 |
0.18 |
| chr15_103334189_103334404 | 0.36 |
Zfp385a |
zinc finger protein 385A |
31 |
0.95 |
| chr1_129272928_129273957 | 0.36 |
Thsd7b |
thrombospondin, type I, domain containing 7B |
21 |
0.98 |
| chr5_114743003_114743280 | 0.36 |
Git2 |
GIT ArfGAP 2 |
727 |
0.5 |
| chr11_95247117_95247412 | 0.36 |
Tac4 |
tachykinin 4 |
14265 |
0.13 |
| chr6_118593411_118594059 | 0.36 |
Cacna1c |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
1691 |
0.37 |
| chr1_172019892_172020043 | 0.35 |
Vangl2 |
VANGL planar cell polarity 2 |
6744 |
0.14 |
| chr5_52604627_52604846 | 0.35 |
8030423F21Rik |
RIKEN cDNA 8030423F21 gene |
14276 |
0.14 |
| chr10_29534276_29534889 | 0.35 |
Gm48159 |
predicted gene, 48159 |
135 |
0.94 |
| chr3_30108418_30108578 | 0.35 |
Mecom |
MDS1 and EVI1 complex locus |
31925 |
0.17 |
| chr8_80057499_80058156 | 0.34 |
Hhip |
Hedgehog-interacting protein |
179 |
0.95 |
| chr7_62420605_62420845 | 0.34 |
Mkrn3 |
makorin, ring finger protein, 3 |
586 |
0.68 |
| chr5_15032326_15032654 | 0.34 |
Gm17019 |
predicted gene 17019 |
508 |
0.84 |
| chr12_8063506_8063657 | 0.33 |
Apob |
apolipoprotein B |
51222 |
0.14 |
| chr6_119479409_119481030 | 0.33 |
Fbxl14 |
F-box and leucine-rich repeat protein 14 |
551 |
0.78 |
| chr10_32889642_32890523 | 0.33 |
Nkain2 |
Na+/K+ transporting ATPase interacting 2 |
234 |
0.95 |
| chr2_105126173_105126893 | 0.33 |
Wt1 |
Wilms tumor 1 homolog |
4 |
0.5 |
| chr16_90283526_90284553 | 0.33 |
Scaf4 |
SR-related CTD-associated factor 4 |
269 |
0.9 |
| chr1_194769289_194769440 | 0.33 |
Plxna2 |
plexin A2 |
4797 |
0.2 |
| chr11_116436453_116437309 | 0.33 |
Ubald2 |
UBA-like domain containing 2 |
2372 |
0.17 |
| chr1_134182287_134182438 | 0.33 |
Chil1 |
chitinase-like 1 |
42 |
0.97 |
| chr13_89371778_89371957 | 0.33 |
Hapln1 |
hyaluronan and proteoglycan link protein 1 |
167929 |
0.04 |
| chr6_99520900_99522395 | 0.33 |
Foxp1 |
forkhead box P1 |
485 |
0.83 |
| chr13_69261120_69261271 | 0.33 |
Gm4812 |
predicted gene 4812 |
84460 |
0.08 |
| chr11_115889200_115889351 | 0.32 |
Myo15b |
myosin XVB |
964 |
0.35 |
| chr5_13470497_13470849 | 0.32 |
Gm43130 |
predicted gene 43130 |
10355 |
0.22 |
| chr5_113492671_113492876 | 0.32 |
Wscd2 |
WSC domain containing 2 |
2021 |
0.38 |
| chr2_28566649_28566800 | 0.32 |
Cel |
carboxyl ester lipase |
3321 |
0.12 |
| chr14_120389109_120389704 | 0.32 |
Gm26679 |
predicted gene, 26679 |
421 |
0.87 |
| chr11_84879754_84880861 | 0.32 |
Myo19 |
myosin XIX |
3 |
0.5 |
| chr13_119741850_119742224 | 0.32 |
Nim1k |
NIM1 serine/threonine protein kinase |
3643 |
0.15 |
| chr5_119743739_119744135 | 0.31 |
Tbx3os2 |
T-box 3, opposite strand 2 |
52719 |
0.1 |
| chr11_74896307_74898160 | 0.31 |
Sgsm2 |
small G protein signaling modulator 2 |
173 |
0.84 |
| chr9_61549545_61549892 | 0.31 |
Gm34424 |
predicted gene, 34424 |
16303 |
0.23 |
| chr12_32995168_32995399 | 0.31 |
Gm24922 |
predicted gene, 24922 |
4727 |
0.16 |
| chr3_135581620_135581778 | 0.31 |
Nfkb1 |
nuclear factor of kappa light polypeptide gene enhancer in B cells 1, p105 |
12742 |
0.16 |
| chr8_105604817_105605984 | 0.31 |
Ripor1 |
RHO family interacting cell polarization regulator 1 |
145 |
0.9 |
| chr4_110152119_110152431 | 0.31 |
Elavl4 |
ELAV like RNA binding protein 4 |
70873 |
0.12 |
| chr2_131040943_131041967 | 0.31 |
Gfra4 |
glial cell line derived neurotrophic factor family receptor alpha 4 |
2 |
0.94 |
| chr7_25729452_25729603 | 0.31 |
Ccdc97 |
coiled-coil domain containing 97 |
10439 |
0.09 |
| chr15_96474728_96474879 | 0.31 |
Scaf11 |
SR-related CTD-associated factor 11 |
13960 |
0.19 |
| chr1_135771295_135771469 | 0.31 |
Phlda3 |
pleckstrin homology like domain, family A, member 3 |
4248 |
0.17 |
| chr1_190042210_190042373 | 0.30 |
Smyd2 |
SET and MYND domain containing 2 |
119928 |
0.05 |
| chr9_56657298_56657483 | 0.30 |
Lingo1 |
leucine rich repeat and Ig domain containing 1 |
9730 |
0.21 |
| chr10_94943541_94945198 | 0.30 |
Plxnc1 |
plexin C1 |
466 |
0.84 |
| chr4_147397309_147397460 | 0.30 |
Gm8178 |
predicted gene 8178 |
18455 |
0.13 |
| chr8_84945228_84946066 | 0.30 |
Rtbdn |
retbindin |
1344 |
0.18 |
| chr5_14978282_14978636 | 0.29 |
Gm10354 |
predicted gene 10354 |
476 |
0.83 |
| chr1_141137215_141137503 | 0.29 |
Gm4845 |
predicted gene 4845 |
119798 |
0.06 |
| chr5_137580251_137580513 | 0.29 |
Tfr2 |
transferrin receptor 2 |
1489 |
0.17 |
| chr11_115831581_115831861 | 0.29 |
Llgl2 |
LLGL2 scribble cell polarity complex component |
247 |
0.85 |
| chr4_136144287_136145200 | 0.29 |
Id3 |
inhibitor of DNA binding 3 |
231 |
0.9 |
| chr15_69107943_69108094 | 0.29 |
4930573C08Rik |
RIKEN cDNA 4930573C08 gene |
126981 |
0.05 |
| chr8_69121372_69122097 | 0.29 |
Atp6v1b2 |
ATPase, H+ transporting, lysosomal V1 subunit B2 |
18606 |
0.14 |
| chr5_137510563_137511218 | 0.29 |
Gm8066 |
predicted gene 8066 |
637 |
0.45 |
| chr6_114131157_114131308 | 0.29 |
Slc6a11 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 11 |
9 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.4 | 2.3 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 0.7 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.2 | 0.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 0.5 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.2 | 0.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 0.5 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.2 | 0.5 | GO:0055118 | negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 | 0.4 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 0.7 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.5 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.2 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.1 | 0.3 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.2 | GO:0072050 | S-shaped body morphogenesis(GO:0072050) |
| 0.1 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.2 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.1 | 0.2 | GO:0001546 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:2001055 | positive regulation of mesenchymal cell apoptotic process(GO:2001055) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.5 | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.0 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.1 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.0 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.0 | 0.1 | GO:0048696 | regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.2 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.2 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.2 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0086045 | membrane depolarization during AV node cell action potential(GO:0086045) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.0 | 0.1 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 1.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.0 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.0 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.0 | 0.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.0 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0060179 | male mating behavior(GO:0060179) |
| 0.0 | 0.1 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.0 | 0.2 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0072235 | distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.0 | 0.1 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.0 | GO:0072034 | renal vesicle induction(GO:0072034) metanephric nephron tubule formation(GO:0072289) |
| 0.0 | 0.1 | GO:0097466 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 1.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:0060751 | branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.1 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0098597 | vocal learning(GO:0042297) imitative learning(GO:0098596) observational learning(GO:0098597) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.0 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.0 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.0 | 0.1 | GO:0070627 | ferrous iron import(GO:0070627) |
| 0.0 | 0.0 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.0 | GO:1903935 | response to sodium arsenite(GO:1903935) |
| 0.0 | 0.1 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) |
| 0.0 | 0.1 | GO:0003163 | sinoatrial node development(GO:0003163) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.0 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.0 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.0 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.2 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 0.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.0 | 0.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.0 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.5 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.0 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.0 | GO:1902805 | positive regulation of synaptic vesicle transport(GO:1902805) |
| 0.0 | 0.1 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.0 | 0.0 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.4 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.1 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:1901142 | insulin metabolic process(GO:1901142) |
| 0.0 | 0.1 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.0 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.0 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0010224 | response to UV-B(GO:0010224) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.1 | GO:1901160 | serotonin metabolic process(GO:0042428) primary amino compound metabolic process(GO:1901160) |
| 0.0 | 0.0 | GO:1902988 | neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.0 | 0.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 1.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.0 | GO:0044860 | protein localization to plasma membrane raft(GO:0044860) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.2 | 3.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.0 | GO:0097452 | GAIT complex(GO:0097452) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.9 | GO:0005167 | neurotrophin TRK receptor binding(GO:0005167) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.3 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.6 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.1 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.2 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0046977 | TAP binding(GO:0046977) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:1990190 | peptide-serine-N-acetyltransferase activity(GO:1990189) peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 2.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0034547 | N-cyclopropylmelamine deaminase activity(GO:0034547) N-cyclopropylammeline deaminase activity(GO:0034548) N-cyclopropylammelide alkylamino hydrolase activity(GO:0034549) 2,5-diamino-6-ribitylamino-4(3H)-pyrimidinone 5'-phosphate deaminase activity(GO:0043723) tRNA-specific adenosine-37 deaminase activity(GO:0043829) archaeal-specific GTP cyclohydrolase activity(GO:0044682) tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.0 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.1 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.0 | 0.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 3.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.0 | REACTOME G1 S TRANSITION | Genes involved in G1/S Transition |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |