Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads
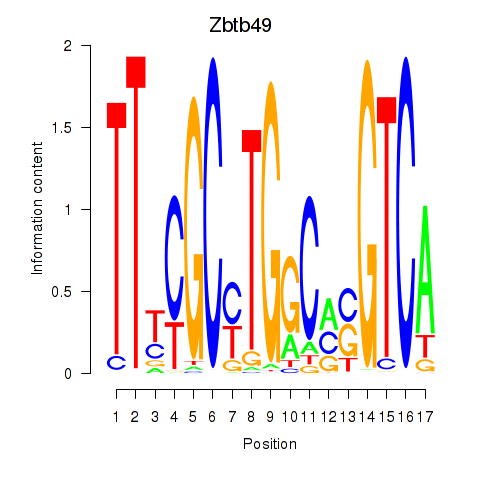
Results for Zbtb49
Z-value: 0.75
Transcription factors associated with Zbtb49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb49
|
ENSMUSG00000029127.9 | zinc finger and BTB domain containing 49 |
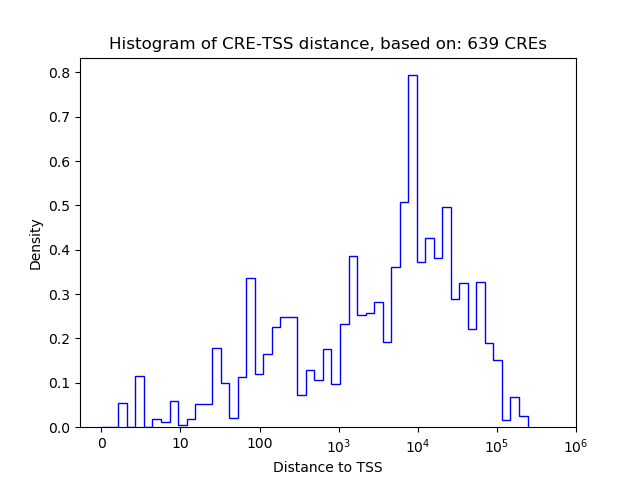
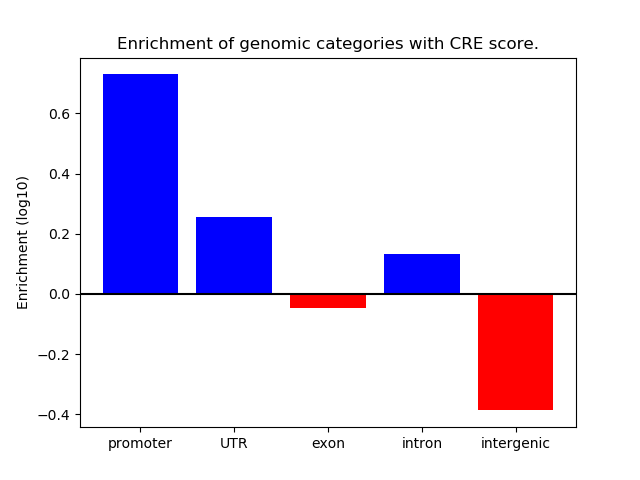
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_38197525_38197734 | Zbtb49 | 6168 | 0.161409 | 0.43 | 1.1e-03 | Click! |
| chr5_38219903_38220668 | Zbtb49 | 128 | 0.577877 | -0.10 | 4.7e-01 | Click! |
Activity of the Zbtb49 motif across conditions
Conditions sorted by the z-value of the Zbtb49 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
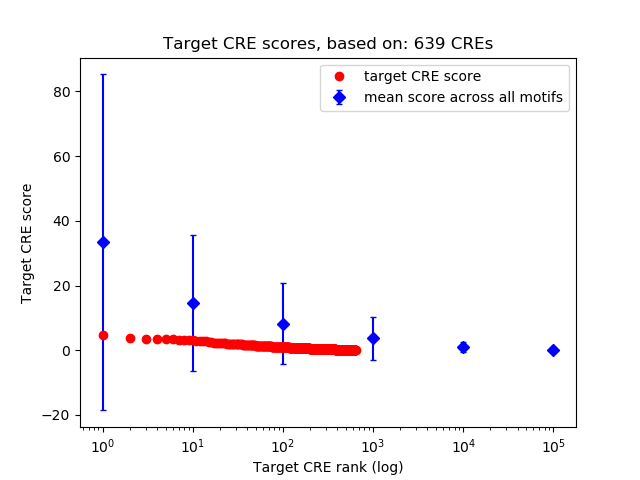
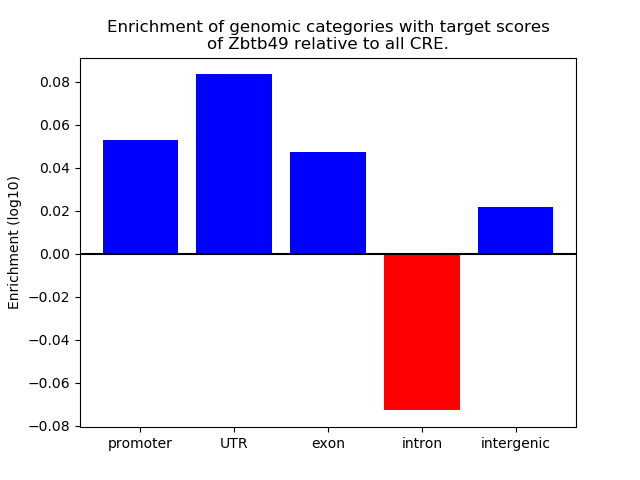
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_99351180_99351446 | 4.73 |
Serpinh1 |
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
1684 |
0.3 |
| chr16_52134647_52134858 | 3.92 |
Cblb |
Casitas B-lineage lymphoma b |
17774 |
0.23 |
| chr5_51899713_51899923 | 3.61 |
Gm42617 |
predicted gene 42617 |
6604 |
0.18 |
| chr1_38835547_38836894 | 3.53 |
Lonrf2 |
LON peptidase N-terminal domain and ring finger 2 |
154 |
0.95 |
| chr2_181156709_181156878 | 3.48 |
Eef1a2 |
eukaryotic translation elongation factor 1 alpha 2 |
221 |
0.88 |
| chr8_4492910_4494136 | 3.37 |
Cers4 |
ceramide synthase 4 |
2 |
0.97 |
| chr11_34348265_34348533 | 3.18 |
Insyn2b |
inhibitory synaptic factor family member 2B |
33577 |
0.16 |
| chr2_49671811_49671963 | 3.16 |
Gm13525 |
predicted gene 13525 |
8612 |
0.24 |
| chr14_34892319_34892570 | 3.14 |
Mir346 |
microRNA 346 |
2165 |
0.38 |
| chr13_84752343_84752734 | 3.09 |
Gm26913 |
predicted gene, 26913 |
61597 |
0.15 |
| chr4_105133429_105133580 | 2.97 |
Prkaa2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
23614 |
0.21 |
| chr1_97848651_97848869 | 2.92 |
Pam |
peptidylglycine alpha-amidating monooxygenase |
7879 |
0.21 |
| chr12_27357279_27357647 | 2.75 |
Sox11 |
SRY (sex determining region Y)-box 11 |
14889 |
0.28 |
| chr17_24736037_24737110 | 2.74 |
Msrb1 |
methionine sulfoxide reductase B1 |
69 |
0.91 |
| chr8_15012305_15012539 | 2.44 |
Kbtbd11 |
kelch repeat and BTB (POZ) domain containing 11 |
1116 |
0.34 |
| chr4_95966590_95967315 | 2.41 |
9530080O11Rik |
RIKEN cDNA 9530080O11 gene |
83 |
0.77 |
| chr19_59043649_59043837 | 2.36 |
Shtn1 |
shootin 1 |
5458 |
0.22 |
| chr4_24430547_24430742 | 2.35 |
Gm27243 |
predicted gene 27243 |
246 |
0.94 |
| chr4_130865591_130865782 | 2.28 |
Gm22660 |
predicted gene, 22660 |
7806 |
0.15 |
| chr18_12570169_12570328 | 2.11 |
Gm22340 |
predicted gene, 22340 |
8289 |
0.17 |
| chr19_7267088_7267830 | 2.09 |
Rcor2 |
REST corepressor 2 |
134 |
0.92 |
| chr2_38132297_38132687 | 2.09 |
Gm44291 |
predicted gene, 44291 |
25196 |
0.17 |
| chr8_124910029_124910183 | 2.08 |
Sprtn |
SprT-like N-terminal domain |
8291 |
0.12 |
| chr1_56726194_56726492 | 2.06 |
Hsfy2 |
heat shock transcription factor, Y-linked 2 |
88908 |
0.09 |
| chr1_109982807_109983363 | 2.05 |
Cdh7 |
cadherin 7, type 2 |
79 |
0.99 |
| chr12_29547825_29548188 | 2.05 |
Myt1l |
myelin transcription factor 1-like |
12784 |
0.23 |
| chr5_114743003_114743280 | 1.94 |
Git2 |
GIT ArfGAP 2 |
727 |
0.5 |
| chr16_22119053_22119204 | 1.84 |
Igf2bp2 |
insulin-like growth factor 2 mRNA binding protein 2 |
2108 |
0.32 |
| chr18_23517169_23517333 | 1.82 |
Dtna |
dystrobrevin alpha |
15508 |
0.28 |
| chr1_132200708_132201234 | 1.81 |
Lemd1 |
LEM domain containing 1 |
8 |
0.96 |
| chr1_83408010_83408453 | 1.78 |
Sphkap |
SPHK1 interactor, AKAP domain containing |
31 |
0.98 |
| chr6_51848127_51848460 | 1.77 |
Skap2 |
src family associated phosphoprotein 2 |
23636 |
0.22 |
| chr1_87175818_87176082 | 1.77 |
Prss56 |
protease, serine 56 |
7363 |
0.1 |
| chr10_84060700_84061314 | 1.77 |
Gm37908 |
predicted gene, 37908 |
751 |
0.67 |
| chr8_110716047_110716632 | 1.77 |
Mtss2 |
MTSS I-BAR domain containing 2 |
5137 |
0.18 |
| chrX_77509141_77509684 | 1.76 |
Tbl1x |
transducin (beta)-like 1 X-linked |
1601 |
0.45 |
| chr12_99474171_99474502 | 1.73 |
Foxn3 |
forkhead box N3 |
24239 |
0.17 |
| chr4_149255354_149255580 | 1.68 |
Kif1b |
kinesin family member 1B |
7488 |
0.18 |
| chr1_43358317_43358468 | 1.67 |
Gm29041 |
predicted gene 29041 |
51192 |
0.12 |
| chr4_150769131_150769447 | 1.60 |
Gm13049 |
predicted gene 13049 |
56444 |
0.1 |
| chr14_34080426_34080774 | 1.58 |
Gm49129 |
predicted gene, 49129 |
4153 |
0.11 |
| chr12_29938165_29938369 | 1.56 |
Pxdn |
peroxidasin |
217 |
0.96 |
| chr4_91244667_91244900 | 1.56 |
Elavl2 |
ELAV like RNA binding protein 1 |
9799 |
0.27 |
| chr6_54507929_54508080 | 1.56 |
Scrn1 |
secernin 1 |
1357 |
0.41 |
| chr8_12105221_12105384 | 1.52 |
B020031H02Rik |
RIKEN cDNA B020031H02 gene |
156963 |
0.03 |
| chr17_56467426_56468397 | 1.52 |
Ptprs |
protein tyrosine phosphatase, receptor type, S |
5695 |
0.16 |
| chr6_18446901_18447365 | 1.47 |
Gm26233 |
predicted gene, 26233 |
2207 |
0.27 |
| chr7_49266538_49266689 | 1.46 |
Nav2 |
neuron navigator 2 |
19768 |
0.21 |
| chr19_10041145_10042475 | 1.46 |
Fads3 |
fatty acid desaturase 3 |
78 |
0.96 |
| chr3_127705467_127706232 | 1.45 |
1500005C15Rik |
RIKEN cDNA 1500005C15 gene |
11178 |
0.11 |
| chr13_24625131_24625340 | 1.44 |
Ripor2 |
RHO family interacting cell polarization regulator 2 |
10608 |
0.19 |
| chrX_69666247_69666398 | 1.41 |
Gm8172 |
predicted pseudogene 8172 |
60077 |
0.15 |
| chr7_96211015_96211192 | 1.39 |
Tenm4 |
teneurin transmembrane protein 4 |
466 |
0.82 |
| chr11_94160820_94161321 | 1.38 |
B230206L02Rik |
RIKEN cDNA B230206L02 gene |
25410 |
0.16 |
| chr18_53294048_53294202 | 1.37 |
Snx24 |
sorting nexing 24 |
48356 |
0.15 |
| chr8_68361979_68362165 | 1.36 |
1700125H03Rik |
RIKEN cDNA 1700125H03 gene |
6362 |
0.22 |
| chr3_38246500_38246797 | 1.35 |
Gm2965 |
predicted gene 2965 |
23197 |
0.18 |
| chr5_111837054_111837274 | 1.35 |
Gm36535 |
predicted gene, 36535 |
43777 |
0.15 |
| chr14_27332479_27332650 | 1.32 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
3502 |
0.27 |
| chr10_44785500_44785699 | 1.31 |
4930431F10Rik |
RIKEN cDNA 4930431F10 gene |
46699 |
0.09 |
| chr4_137476332_137476648 | 1.31 |
Hspg2 |
perlecan (heparan sulfate proteoglycan 2) |
7687 |
0.14 |
| chr12_92152419_92152618 | 1.29 |
Gm40598 |
predicted gene, 40598 |
8921 |
0.26 |
| chr10_61080137_61080290 | 1.29 |
Pcbd1 |
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 |
9118 |
0.13 |
| chr14_7818766_7819099 | 1.29 |
Flnb |
filamin, beta |
975 |
0.49 |
| chr8_95703143_95704225 | 1.27 |
Ndrg4 |
N-myc downstream regulated gene 4 |
614 |
0.57 |
| chr10_39266683_39266865 | 1.24 |
Gm31562 |
predicted gene, 31562 |
21728 |
0.17 |
| chr2_106194434_106194642 | 1.23 |
Dcdc5 |
doublecortin domain containing 5 |
27519 |
0.21 |
| chr13_34156890_34157101 | 1.20 |
Psmg4 |
proteasome (prosome, macropain) assembly chaperone 4 |
5969 |
0.13 |
| chr4_108779892_108781170 | 1.20 |
Zfyve9 |
zinc finger, FYVE domain containing 9 |
28 |
0.97 |
| chr5_38813852_38814038 | 1.20 |
Clnk |
cytokine-dependent hematopoietic cell linker |
62867 |
0.12 |
| chr2_165043149_165043683 | 1.18 |
Ncoa5 |
nuclear receptor coactivator 5 |
8549 |
0.14 |
| chr1_52454422_52454927 | 1.18 |
Gm23460 |
predicted gene, 23460 |
10130 |
0.16 |
| chr5_123498102_123498542 | 1.17 |
Il31 |
interleukin 31 |
8833 |
0.07 |
| chr4_91376156_91376714 | 1.15 |
Elavl2 |
ELAV like RNA binding protein 1 |
1 |
0.98 |
| chr5_35280918_35281069 | 1.15 |
Adra2c |
adrenergic receptor, alpha 2c |
2674 |
0.31 |
| chr14_22679666_22680655 | 1.14 |
Lrmda |
leucine rich melanocyte differentiation associated |
83647 |
0.09 |
| chr2_152093402_152093892 | 1.13 |
Srxn1 |
sulfiredoxin 1 homolog (S. cerevisiae) |
11869 |
0.13 |
| chr12_15800918_15801338 | 1.12 |
Mir6387 |
microRNA 6387 |
27 |
0.97 |
| chr7_49430942_49431093 | 1.12 |
Nav2 |
neuron navigator 2 |
19743 |
0.21 |
| chr16_94467495_94467646 | 1.08 |
Ttc3 |
tetratricopeptide repeat domain 3 |
1048 |
0.48 |
| chr9_21037477_21037931 | 1.07 |
Gm26592 |
predicted gene, 26592 |
78 |
0.91 |
| chr9_46599951_46600255 | 1.07 |
Gm48945 |
predicted gene, 48945 |
7152 |
0.17 |
| chr4_130519822_130520506 | 1.06 |
Nkain1 |
Na+/K+ transporting ATPase interacting 1 |
37824 |
0.17 |
| chr6_22834709_22835019 | 1.05 |
Gm43629 |
predicted gene 43629 |
29535 |
0.14 |
| chr10_117042279_117042645 | 1.04 |
Gm10747 |
predicted gene 10747 |
1284 |
0.32 |
| chr8_126667748_126668147 | 1.04 |
Irf2bp2 |
interferon regulatory factor 2 binding protein 2 |
73961 |
0.1 |
| chr1_8863954_8864105 | 1.04 |
Gm7445 |
predicted gene 7445 |
7266 |
0.17 |
| chr10_7323409_7323560 | 1.02 |
Gm21321 |
predicted gene, 21321 |
67299 |
0.11 |
| chr6_39787853_39788004 | 1.01 |
Mrps33 |
mitochondrial ribosomal protein S33 |
18114 |
0.16 |
| chr1_186555717_186555886 | 1.00 |
A730004F24Rik |
RIKEN cDNA A730004F24 gene |
2671 |
0.35 |
| chr9_91378183_91378872 | 1.00 |
Zic4 |
zinc finger protein of the cerebellum 4 |
115 |
0.94 |
| chr19_5770648_5771783 | 0.99 |
Scyl1 |
SCY1-like 1 (S. cerevisiae) |
38 |
0.93 |
| chr5_149523996_149524147 | 0.99 |
Wdr95 |
WD40 repeat domain 95 |
4608 |
0.16 |
| chr3_93445265_93445429 | 0.99 |
Tchh |
trichohyalin |
3017 |
0.12 |
| chr15_102257553_102258012 | 0.98 |
Rarg |
retinoic acid receptor, gamma |
265 |
0.82 |
| chr19_25598711_25598994 | 0.98 |
Dmrt3 |
doublesex and mab-3 related transcription factor 3 |
11449 |
0.22 |
| chr1_42851025_42851764 | 0.98 |
Mrps9 |
mitochondrial ribosomal protein S9 |
109 |
0.97 |
| chr11_101732407_101733143 | 0.97 |
Dhx8 |
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
144 |
0.94 |
| chr17_15247926_15248133 | 0.97 |
Gm5091 |
predicted gene 5091 |
112 |
0.97 |
| chr2_115895484_115896072 | 0.96 |
Meis2 |
Meis homeobox 2 |
22816 |
0.26 |
| chr6_84143462_84143613 | 0.94 |
Dysf |
dysferlin |
3210 |
0.27 |
| chr3_102734319_102735029 | 0.94 |
Tspan2 |
tetraspanin 2 |
145 |
0.87 |
| chr5_124211305_124211583 | 0.93 |
Pitpnm2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
4974 |
0.13 |
| chr4_148384621_148385111 | 0.91 |
Gm13200 |
predicted gene 13200 |
25406 |
0.13 |
| chr7_64156624_64156775 | 0.90 |
Trpm1 |
transient receptor potential cation channel, subfamily M, member 1 |
2771 |
0.23 |
| chr5_25760091_25760623 | 0.90 |
Actr3b |
ARP3 actin-related protein 3B |
318 |
0.89 |
| chr2_91956953_91957140 | 0.88 |
Dgkz |
diacylglycerol kinase zeta |
6611 |
0.12 |
| chr10_4179226_4179383 | 0.88 |
Gm25369 |
predicted gene, 25369 |
9417 |
0.21 |
| chr6_30079051_30079222 | 0.87 |
Nrf1 |
nuclear respiratory factor 1 |
2815 |
0.25 |
| chr1_87184907_87185090 | 0.86 |
Prss56 |
protease, serine 56 |
1685 |
0.21 |
| chr15_68230867_68231032 | 0.86 |
Zfat |
zinc finger and AT hook domain containing |
27838 |
0.18 |
| chr5_81021312_81021925 | 0.86 |
Adgrl3 |
adhesion G protein-coupled receptor L3 |
25 |
0.99 |
| chr3_88538384_88539050 | 0.86 |
Mir1905 |
microRNA 1905 |
2335 |
0.12 |
| chr3_4798553_4798746 | 0.86 |
1110015O18Rik |
RIKEN cDNA 1110015O18 gene |
59 |
0.98 |
| chr4_58184913_58185294 | 0.85 |
Svep1 |
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
21493 |
0.25 |
| chr13_52283378_52283752 | 0.85 |
Gm48199 |
predicted gene, 48199 |
103154 |
0.07 |
| chr17_26658958_26659109 | 0.84 |
Atp6v0e |
ATPase, H+ transporting, lysosomal V0 subunit E |
4198 |
0.16 |
| chr14_54572707_54573548 | 0.83 |
Ajuba |
ajuba LIM protein |
1045 |
0.3 |
| chr13_56703202_56704180 | 0.81 |
Smad5 |
SMAD family member 5 |
23 |
0.98 |
| chr5_35448262_35448469 | 0.81 |
Gm43377 |
predicted gene 43377 |
52267 |
0.09 |
| chr13_117220464_117220939 | 0.81 |
Emb |
embigin |
15 |
0.86 |
| chr12_113138507_113138658 | 0.80 |
Crip2 |
cysteine rich protein 2 |
1654 |
0.22 |
| chr9_23120943_23121094 | 0.80 |
Bmper |
BMP-binding endothelial regulator |
102058 |
0.08 |
| chr15_85724908_85725104 | 0.80 |
Ppara |
peroxisome proliferator activated receptor alpha |
9977 |
0.13 |
| chr7_128687903_128688560 | 0.80 |
Inpp5f |
inositol polyphosphate-5-phosphatase F |
258 |
0.51 |
| chr7_18858684_18858835 | 0.79 |
Pglyrp1 |
peptidoglycan recognition protein 1 |
12572 |
0.09 |
| chr9_30149631_30149782 | 0.78 |
Gm47677 |
predicted gene, 47677 |
17869 |
0.26 |
| chr4_63291492_63291785 | 0.77 |
Col27a1 |
collagen, type XXVII, alpha 1 |
8125 |
0.16 |
| chr4_82344790_82345141 | 0.77 |
n-R5s188 |
nuclear encoded rRNA 5S 188 |
94445 |
0.08 |
| chr2_70126801_70127190 | 0.77 |
Myo3b |
myosin IIIB |
30697 |
0.2 |
| chr15_27503321_27503541 | 0.77 |
B230362B09Rik |
RIKEN cDNA B230362B09 gene |
481 |
0.78 |
| chr12_73045160_73045311 | 0.77 |
Six1 |
sine oculis-related homeobox 1 |
56 |
0.98 |
| chr12_91066386_91066537 | 0.76 |
Cep128 |
centrosomal protein 128 |
3050 |
0.24 |
| chr7_99304833_99305309 | 0.76 |
Gm45591 |
predicted gene 45591 |
16129 |
0.12 |
| chr9_27568724_27568875 | 0.76 |
Gm18585 |
predicted gene, 18585 |
14593 |
0.18 |
| chr10_41594987_41595356 | 0.75 |
Ccdc162 |
coiled-coil domain containing 162 |
7418 |
0.15 |
| chr6_119674280_119674487 | 0.74 |
Erc1 |
ELKS/RAB6-interacting/CAST family member 1 |
52278 |
0.15 |
| chr6_30698043_30698194 | 0.74 |
Cep41 |
centrosomal protein 41 |
4369 |
0.14 |
| chr1_128758816_128759186 | 0.73 |
Gm29599 |
predicted gene 29599 |
8075 |
0.23 |
| chr8_12430018_12430574 | 0.73 |
Gm25239 |
predicted gene, 25239 |
33893 |
0.1 |
| chr7_79578006_79578675 | 0.73 |
Gm45168 |
predicted gene 45168 |
728 |
0.49 |
| chr13_115002286_115002437 | 0.73 |
Gm47776 |
predicted gene, 47776 |
67293 |
0.09 |
| chr4_126262506_126262833 | 0.73 |
Trappc3 |
trafficking protein particle complex 3 |
269 |
0.86 |
| chr2_160766567_160766718 | 0.72 |
Plcg1 |
phospholipase C, gamma 1 |
6524 |
0.18 |
| chr8_25976584_25977311 | 0.72 |
Hgsnat |
heparan-alpha-glucosaminide N-acetyltransferase |
194 |
0.91 |
| chr12_27151151_27151367 | 0.72 |
Gm9866 |
predicted gene 9866 |
9052 |
0.3 |
| chr11_70200228_70200379 | 0.71 |
Slc16a11 |
solute carrier family 16 (monocarboxylic acid transporters), member 11 |
12449 |
0.07 |
| chr7_125764515_125764737 | 0.71 |
D430042O09Rik |
RIKEN cDNA D430042O09 gene |
21629 |
0.21 |
| chr7_16203170_16203481 | 0.71 |
Dhx34 |
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
876 |
0.44 |
| chr14_99944188_99944685 | 0.70 |
Klf12 |
Kruppel-like factor 12 |
728 |
0.78 |
| chr13_51155566_51155768 | 0.70 |
Nxnl2 |
nucleoredoxin-like 2 |
15355 |
0.16 |
| chr16_7447825_7447988 | 0.70 |
Rbfox1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
94936 |
0.1 |
| chr4_45422743_45422894 | 0.70 |
Slc25a51 |
solute carrier family 25, member 51 |
14052 |
0.15 |
| chr3_120982360_120982511 | 0.68 |
Gm43444 |
predicted gene 43444 |
38341 |
0.16 |
| chr16_4643666_4644188 | 0.68 |
Coro7 |
coronin 7 |
2789 |
0.15 |
| chr16_21370010_21370180 | 0.68 |
Magef1 |
melanoma antigen family F, 1 |
36739 |
0.15 |
| chr11_87005447_87005718 | 0.68 |
Gm22387 |
predicted gene, 22387 |
8319 |
0.15 |
| chr7_118991798_118991949 | 0.68 |
Gprc5b |
G protein-coupled receptor, family C, group 5, member B |
3338 |
0.25 |
| chr2_39252305_39252456 | 0.66 |
Gm13504 |
predicted gene 13504 |
14939 |
0.13 |
| chr7_16131212_16131667 | 0.66 |
Slc8a2 |
solute carrier family 8 (sodium/calcium exchanger), member 2 |
1091 |
0.37 |
| chr2_157487979_157488189 | 0.66 |
Src |
Rous sarcoma oncogene |
30999 |
0.11 |
| chr19_48333070_48333382 | 0.65 |
Gm23857 |
predicted gene, 23857 |
92581 |
0.08 |
| chr10_108364964_108365547 | 0.65 |
Gm23105 |
predicted gene, 23105 |
1590 |
0.4 |
| chr10_91881912_91882107 | 0.65 |
Gm31592 |
predicted gene, 31592 |
6820 |
0.31 |
| chr15_99528118_99528269 | 0.64 |
Faim2 |
Fas apoptotic inhibitory molecule 2 |
28 |
0.96 |
| chr18_60710262_60710458 | 0.64 |
Ndst1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
3029 |
0.23 |
| chr5_38560766_38562198 | 0.64 |
Wdr1 |
WD repeat domain 1 |
61 |
0.97 |
| chr17_79231357_79231508 | 0.64 |
Gm5230 |
predicted gene 5230 |
26617 |
0.21 |
| chr11_50377311_50377861 | 0.62 |
Hnrnph1 |
heterogeneous nuclear ribonucleoprotein H1 |
133 |
0.94 |
| chr9_21130693_21131801 | 0.62 |
Tyk2 |
tyrosine kinase 2 |
4 |
0.95 |
| chr5_24685391_24686443 | 0.61 |
Nub1 |
negative regulator of ubiquitin-like proteins 1 |
70 |
0.96 |
| chr1_32172857_32173290 | 0.61 |
Khdrbs2 |
KH domain containing, RNA binding, signal transduction associated 2 |
186 |
0.97 |
| chr1_172221317_172221900 | 0.61 |
Casq1 |
calsequestrin 1 |
1740 |
0.2 |
| chr1_193989994_193990577 | 0.61 |
Gm21362 |
predicted gene, 21362 |
123276 |
0.06 |
| chr1_119421277_119422204 | 0.60 |
Inhbb |
inhibin beta-B |
508 |
0.79 |
| chr13_52223178_52223329 | 0.60 |
Gm48199 |
predicted gene, 48199 |
42842 |
0.19 |
| chr3_105052490_105052858 | 0.59 |
Cttnbp2nl |
CTTNBP2 N-terminal like |
251 |
0.92 |
| chr11_98959522_98960759 | 0.59 |
Rara |
retinoic acid receptor, alpha |
272 |
0.84 |
| chr12_105336106_105336602 | 0.59 |
Tunar |
Tcl1 upstream neural differentiation associated RNA |
240 |
0.93 |
| chr3_106684799_106685270 | 0.59 |
Lrif1 |
ligand dependent nuclear receptor interacting factor 1 |
36 |
0.98 |
| chr9_37090365_37090550 | 0.58 |
Pknox2 |
Pbx/knotted 1 homeobox 2 |
7148 |
0.16 |
| chr7_140063115_140063403 | 0.58 |
Msx3 |
msh homeobox 3 |
14170 |
0.08 |
| chr5_90223854_90224505 | 0.58 |
Cox18 |
cytochrome c oxidase assembly protein 18 |
178 |
0.96 |
| chr14_64125810_64126264 | 0.57 |
9630015K15Rik |
RIKEN cDNA 9630015K15 gene |
9723 |
0.13 |
| chr1_13189266_13189438 | 0.57 |
Gm38376 |
predicted gene, 38376 |
9638 |
0.16 |
| chr2_155299641_155299792 | 0.56 |
Pigu |
phosphatidylinositol glycan anchor biosynthesis, class U |
1586 |
0.33 |
| chr1_164454802_164455037 | 0.56 |
Atp1b1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
1142 |
0.41 |
| chr17_83187838_83188005 | 0.56 |
Pkdcc |
protein kinase domain containing, cytoplasmic |
27371 |
0.19 |
| chr10_83722468_83723424 | 0.56 |
1500009L16Rik |
RIKEN cDNA 1500009L16 gene |
20 |
0.98 |
| chr1_78167977_78168321 | 0.56 |
Pax3 |
paired box 3 |
28689 |
0.2 |
| chr8_88939205_88939356 | 0.55 |
Gm45504 |
predicted gene 45504 |
76206 |
0.09 |
| chr1_134026827_134026978 | 0.55 |
Gm18970 |
predicted gene, 18970 |
6476 |
0.15 |
| chr9_20830924_20831267 | 0.55 |
AC170598.1 |
complement component 3 precursor (C3P1) pseudogene |
8433 |
0.1 |
| chr12_58214120_58214334 | 0.54 |
Sstr1 |
somatostatin receptor 1 |
2423 |
0.4 |
| chr18_69384579_69384730 | 0.54 |
Tcf4 |
transcription factor 4 |
31243 |
0.22 |
| chr10_62651133_62651769 | 0.54 |
Ddx50 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
233 |
0.88 |
| chr1_190979223_190979743 | 0.54 |
Vash2 |
vasohibin 2 |
187 |
0.9 |
| chr12_78077415_78077566 | 0.54 |
Gm24994 |
predicted gene, 24994 |
88056 |
0.08 |
| chr1_170110460_170110694 | 0.53 |
Ddr2 |
discoidin domain receptor family, member 2 |
16 |
0.98 |
| chr5_46201957_46202226 | 0.52 |
Gm7931 |
predicted pseudogene 7931 |
212253 |
0.02 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0030091 | protein repair(GO:0030091) |
| 0.5 | 2.4 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.4 | 1.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.3 | 0.8 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 0.9 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 0.8 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.2 | 0.8 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 0.6 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.2 | 0.5 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.5 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.5 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 0.6 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 3.1 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.1 | 0.6 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 | 0.8 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 0.4 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.1 | 0.5 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.1 | 0.3 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.5 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.1 | 0.4 | GO:0071883 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.1 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.2 | GO:1901874 | regulation of post-translational protein modification(GO:1901873) negative regulation of post-translational protein modification(GO:1901874) |
| 0.1 | 0.4 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 1.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.4 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.1 | 0.6 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 0.3 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.1 | 0.9 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.1 | 0.9 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.6 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.8 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 3.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.4 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.2 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.5 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.1 | 0.6 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.2 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 0.1 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 | 0.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 1.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0046499 | S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.6 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 0.0 | 0.2 | GO:1904587 | glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 | 0.2 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.7 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.2 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.8 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.2 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.4 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) regulation of membrane lipid metabolic process(GO:1905038) |
| 0.0 | 0.0 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:0048505 | regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.1 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.0 | 0.2 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.4 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.3 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0060331 | negative regulation of response to interferon-gamma(GO:0060331) negative regulation of interferon-gamma-mediated signaling pathway(GO:0060336) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.2 | GO:1904871 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) regulation of ubiquitin-specific protease activity(GO:2000152) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:0046836 | glycolipid transport(GO:0046836) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.0 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0032827 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.7 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0046337 | phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.0 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:1990123 | L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.0 | 0.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.0 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.0 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.0 | GO:0021626 | hindbrain maturation(GO:0021578) pons maturation(GO:0021586) central nervous system maturation(GO:0021626) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.0 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) |
| 0.0 | 0.1 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.0 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.0 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0072309 | mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) |
| 0.0 | 0.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.0 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.0 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.3 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.0 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.0 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0060283 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.0 | 0.0 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.0 | 0.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.0 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.0 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.0 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.0 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.0 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.5 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.2 | GO:0035838 | growing cell tip(GO:0035838) |
| 0.1 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 1.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 1.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 1.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 1.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 2.5 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.2 | 0.6 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 1.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.2 | 0.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 0.9 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 0.5 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.2 | 3.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.3 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.6 | GO:0038064 | collagen receptor activity(GO:0038064) |
| 0.1 | 0.4 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.1 | 0.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.6 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.1 | 0.4 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 0.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.1 | 0.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.0 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 1.3 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.8 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0034842 | 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 1.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.4 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.0 | 0.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0046428 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 1.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 2.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.0 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.0 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.0 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.0 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0043883 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.0 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0008796 | bis(5'-nucleosyl)-tetraphosphatase activity(GO:0008796) |
| 0.0 | 0.0 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.1 | REACTOME MITOTIC G2 G2 M PHASES | Genes involved in Mitotic G2-G2/M phases |
| 0.1 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 1.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.0 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |