Project
ENCODE: ATAC-seq of different tissues during embryonic development
Navigation
Downloads

CREMA is a free online tool that recognizes most
important transcription factors that change the chromatin
state across different samples.
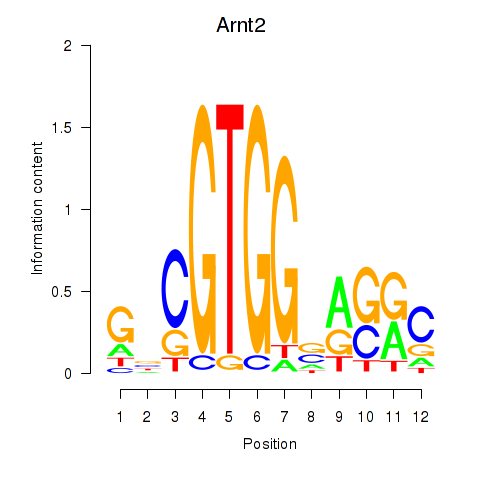
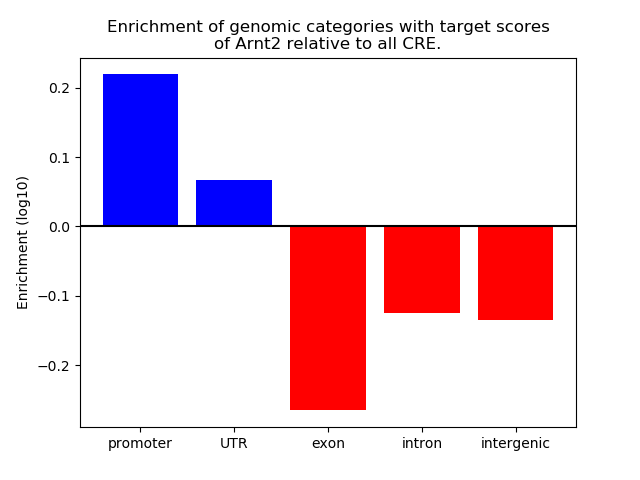
Results for Arnt2
Z-value: 0.41
Transcription factors associated with Arnt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Arnt2
|
ENSMUSG00000015709.8 | Arnt2 |
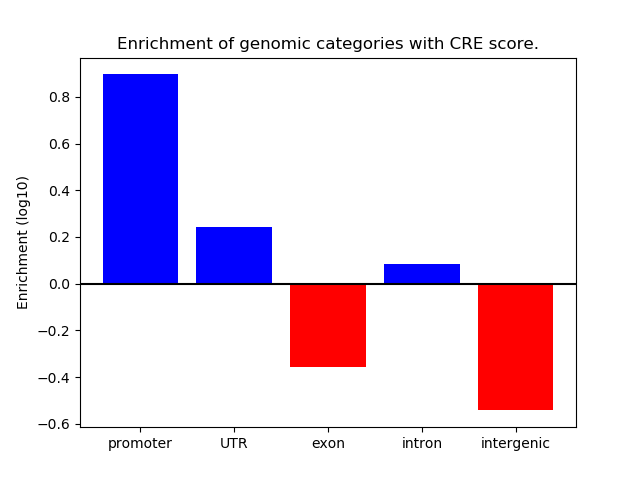
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| Arnt2 | chr7_84372464_84372615 | 8 | 0.976165 | -0.40 | 2.8e-03 | Click! |
| Arnt2 | chr7_84275389_84275540 | 407 | 0.831638 | 0.34 | 1.1e-02 | Click! |
| Arnt2 | chr7_84285970_84286121 | 3651 | 0.209795 | -0.33 | 1.5e-02 | Click! |
| Arnt2 | chr7_84403485_84403681 | 6300 | 0.181922 | -0.30 | 2.4e-02 | Click! |
| Arnt2 | chr7_84307654_84307850 | 3226 | 0.213132 | -0.30 | 2.8e-02 | Click! |
Activity of the Arnt2 motif across conditions
Conditions sorted by the z-value of the Arnt2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
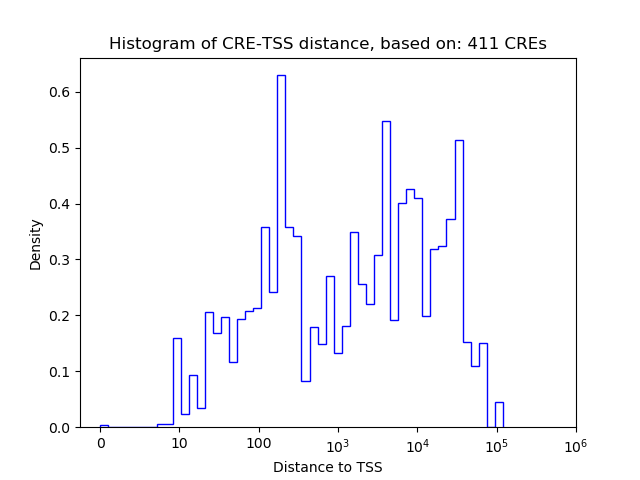
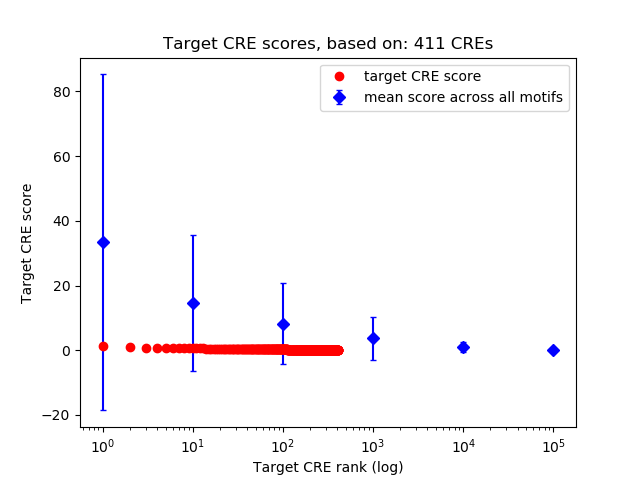
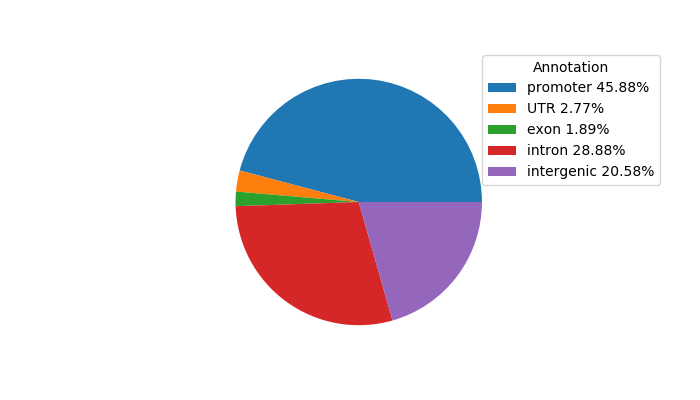
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_15253127_15253318 | 1.36 |
Gm5404 |
predicted gene 5404 |
34378 |
0.16 |
| chr2_170130477_170131578 | 0.90 |
Zfp217 |
zinc finger protein 217 |
193 |
0.97 |
| chr12_69437182_69437482 | 0.81 |
5830428M24Rik |
RIKEN cDNA 5830428M24 gene |
30060 |
0.12 |
| chr1_85251281_85252082 | 0.77 |
C130026I21Rik |
RIKEN cDNA C130026I21 gene |
2867 |
0.17 |
| chr14_115040506_115042372 | 0.75 |
Mir17hg |
Mir17 host gene (non-protein coding) |
1440 |
0.19 |
| chr15_97042032_97042236 | 0.73 |
Slc38a4 |
solute carrier family 38, member 4 |
7953 |
0.28 |
| chr2_152636359_152637068 | 0.67 |
Rem1 |
rad and gem related GTP binding protein 1 |
3746 |
0.1 |
| chr19_23758061_23759360 | 0.65 |
Apba1 |
amyloid beta (A4) precursor protein binding, family A, member 1 |
177 |
0.94 |
| chr1_69106914_69108095 | 0.64 |
Erbb4 |
erb-b2 receptor tyrosine kinase 4 |
252 |
0.82 |
| chr1_85166981_85167344 | 0.57 |
Gm6264 |
predicted gene 6264 |
6299 |
0.11 |
| chr7_71960732_71961203 | 0.55 |
1700011C11Rik |
RIKEN cDNA 1700011C11 gene |
206 |
0.9 |
| chr11_36873121_36873272 | 0.55 |
Tenm2 |
teneurin transmembrane protein 2 |
70970 |
0.14 |
| chr11_96414118_96414269 | 0.54 |
Gm11531 |
predicted gene 11531 |
22081 |
0.1 |
| chr13_72419489_72419940 | 0.52 |
Rpl9-ps4 |
ribosomal protein L9, pseudogene 4 |
35626 |
0.14 |
| chr13_73263734_73263998 | 0.50 |
Irx4 |
Iroquois homeobox 4 |
3369 |
0.2 |
| chr7_44553417_44554082 | 0.49 |
Nr1h2 |
nuclear receptor subfamily 1, group H, member 2 |
41 |
0.92 |
| chr1_85092888_85093459 | 0.48 |
Gm10553 |
predicted gene 10553 |
6339 |
0.1 |
| chr14_19131787_19131938 | 0.47 |
Gm8582 |
predicted gene 8582 |
4062 |
0.15 |
| chr8_122511564_122511947 | 0.47 |
Gm26497 |
predicted gene, 26497 |
8919 |
0.09 |
| chr7_118491741_118492257 | 0.47 |
Itpripl2 |
inositol 1,4,5-triphosphate receptor interacting protein-like 2 |
24 |
0.97 |
| chr2_101752020_101752171 | 0.46 |
Prr5l |
proline rich 5 like |
45555 |
0.14 |
| chr11_85832197_85833021 | 0.45 |
Tbx2 |
T-box 2 |
58 |
0.84 |
| chr9_18473066_18474201 | 0.42 |
Zfp558 |
zinc finger protein 558 |
74 |
0.95 |
| chr19_43957421_43957601 | 0.42 |
Cpn1 |
carboxypeptidase N, polypeptide 1 |
11182 |
0.13 |
| chr19_5094164_5095202 | 0.42 |
Cnih2 |
cornichon family AMPA receptor auxiliary protein 2 |
3699 |
0.08 |
| chr19_38026271_38026463 | 0.41 |
Myof |
myoferlin |
16984 |
0.15 |
| chr8_121786646_121786834 | 0.41 |
Klhdc4 |
kelch domain containing 4 |
10688 |
0.12 |
| chr10_80948620_80948940 | 0.41 |
Gm3828 |
predicted gene 3828 |
5930 |
0.1 |
| chr17_25433917_25434633 | 0.40 |
Cacna1h |
calcium channel, voltage-dependent, T type, alpha 1H subunit |
492 |
0.69 |
| chr2_127415922_127416073 | 0.39 |
Gpat2 |
glycerol-3-phosphate acyltransferase 2, mitochondrial |
9202 |
0.15 |
| chr1_191184878_191185214 | 0.38 |
Atf3 |
activating transcription factor 3 |
1706 |
0.28 |
| chr1_85617918_85618139 | 0.38 |
Gm10552 |
predicted gene 10552 |
6132 |
0.1 |
| chr7_63816080_63816231 | 0.38 |
Gm7482 |
predicted gene 7482 |
4825 |
0.18 |
| chr12_84408114_84409175 | 0.37 |
Entpd5 |
ectonucleoside triphosphate diphosphohydrolase 5 |
340 |
0.53 |
| chr12_86882228_86883612 | 0.37 |
Irf2bpl |
interferon regulatory factor 2 binding protein-like |
1878 |
0.33 |
| chr15_8279549_8279700 | 0.37 |
Cplane1 |
ciliogenesis and planar polarity effector 1 |
32521 |
0.18 |
| chr11_74896307_74898160 | 0.36 |
Sgsm2 |
small G protein signaling modulator 2 |
173 |
0.84 |
| chr2_26604241_26604792 | 0.35 |
Agpat2 |
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta) |
99 |
0.92 |
| chr7_142561215_142561756 | 0.35 |
Nctc1 |
non-coding transcript 1 |
2889 |
0.14 |
| chr17_75014632_75015003 | 0.35 |
Ltbp1 |
latent transforming growth factor beta binding protein 1 |
8523 |
0.24 |
| chr16_38362984_38363227 | 0.35 |
Popdc2 |
popeye domain containing 2 |
860 |
0.48 |
| chr2_26503184_26504033 | 0.35 |
Notch1 |
notch 1 |
214 |
0.89 |
| chr2_138256185_138257474 | 0.34 |
Btbd3 |
BTB (POZ) domain containing 3 |
159 |
0.98 |
| chr3_96451263_96451652 | 0.34 |
BC107364 |
cDNA sequence BC107364 |
849 |
0.24 |
| chr2_74732641_74732796 | 0.34 |
Hoxd3 |
homeobox D3 |
195 |
0.81 |
| chr7_99351180_99351446 | 0.34 |
Serpinh1 |
serine (or cysteine) peptidase inhibitor, clade H, member 1 |
1684 |
0.3 |
| chr10_86300212_86300618 | 0.34 |
Timp3 |
tissue inhibitor of metalloproteinase 3 |
43 |
0.98 |
| chr3_8717321_8717847 | 0.34 |
C230057A21Rik |
RIKEN cDNA C230057A21 gene |
18616 |
0.14 |
| chr14_27114666_27114961 | 0.34 |
Arhgef3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
86 |
0.98 |
| chr11_81612309_81613487 | 0.33 |
Gm11418 |
predicted gene 11418 |
25537 |
0.22 |
| chr9_32901255_32901639 | 0.33 |
Gm27162 |
predicted gene 27162 |
27519 |
0.18 |
| chr1_39721483_39721634 | 0.33 |
Rfx8 |
regulatory factor X 8 |
561 |
0.78 |
| chr10_103001022_103001290 | 0.33 |
Alx1 |
ALX homeobox 1 |
21418 |
0.19 |
| chr5_128598799_128598977 | 0.33 |
Gm42498 |
predicted gene 42498 |
932 |
0.43 |
| chr7_100911356_100911541 | 0.32 |
Arhgef17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
16758 |
0.13 |
| chr15_66961775_66961932 | 0.32 |
Ndrg1 |
N-myc downstream regulated gene 1 |
6072 |
0.18 |
| chr10_121475538_121476618 | 0.32 |
Rassf3 |
Ras association (RalGDS/AF-6) domain family member 3 |
269 |
0.86 |
| chr5_125264626_125264993 | 0.32 |
Gm32585 |
predicted gene, 32585 |
8006 |
0.18 |
| chr5_107195880_107196420 | 0.32 |
Tgfbr3 |
transforming growth factor, beta receptor III |
3783 |
0.2 |
| chr7_110982411_110982739 | 0.32 |
Mrvi1 |
MRV integration site 1 |
114 |
0.97 |
| chr8_11476357_11476528 | 0.31 |
E230013L22Rik |
RIKEN cDNA E230013L22 gene |
1487 |
0.25 |
| chr2_72614373_72614524 | 0.31 |
Pex13-ps |
peroxisomal biogenesis factor 13, pseudogene |
17822 |
0.16 |
| chr1_88213749_88214214 | 0.31 |
Ugt1a1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 |
2022 |
0.13 |
| chr14_27823142_27823309 | 0.31 |
Erc2 |
ELKS/RAB6-interacting/CAST family member 2 |
24667 |
0.22 |
| chr8_15027100_15028701 | 0.31 |
Gm37844 |
predicted gene, 37844 |
522 |
0.44 |
| chr9_50752051_50752676 | 0.30 |
Hspb2 |
heat shock protein 2 |
9 |
0.88 |
| chr5_43232754_43234178 | 0.30 |
Cpeb2 |
cytoplasmic polyadenylation element binding protein 2 |
296 |
0.85 |
| chr1_132346162_132346444 | 0.30 |
F730311O21Rik |
RIKEN cDNA F730311O21 gene |
4280 |
0.14 |
| chr13_35740610_35741587 | 0.30 |
Cdyl |
chromodomain protein, Y chromosome-like |
215 |
0.94 |
| chr4_135876235_135877013 | 0.30 |
Pnrc2 |
proline-rich nuclear receptor coactivator 2 |
2774 |
0.15 |
| chr13_41605449_41606727 | 0.29 |
Tmem170b |
transmembrane protein 170B |
128 |
0.96 |
| chr17_26414662_26415402 | 0.29 |
Neurl1b |
neuralized E3 ubiquitin protein ligase 1B |
67 |
0.97 |
| chr4_149954471_149955779 | 0.29 |
Spsb1 |
splA/ryanodine receptor domain and SOCS box containing 1 |
82 |
0.97 |
| chr6_63257322_63257806 | 0.29 |
9330118I20Rik |
RIKEN cDNA 9330118I20 gene |
61 |
0.93 |
| chr8_69995796_69997172 | 0.27 |
Gatad2a |
GATA zinc finger domain containing 2A |
100 |
0.95 |
| chr7_45588766_45589660 | 0.27 |
Bcat2 |
branched chain aminotransferase 2, mitochondrial |
3792 |
0.07 |
| chr5_140606846_140607467 | 0.27 |
Lfng |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
164 |
0.83 |
| chr15_81585272_81586453 | 0.27 |
Gm23880 |
predicted gene, 23880 |
271 |
0.52 |
| chr5_53998183_53999428 | 0.27 |
Stim2 |
stromal interaction molecule 2 |
240 |
0.95 |
| chr1_89538633_89538887 | 0.27 |
Gm25180 |
predicted gene, 25180 |
24600 |
0.16 |
| chr5_147758193_147758351 | 0.27 |
Gm43156 |
predicted gene 43156 |
12756 |
0.19 |
| chr1_72945188_72945345 | 0.27 |
1700027A15Rik |
RIKEN cDNA 1700027A15 gene |
39578 |
0.16 |
| chr2_26595840_26596165 | 0.27 |
Egfl7 |
EGF-like domain 7 |
3855 |
0.1 |
| chr13_56588245_56589118 | 0.26 |
2010203P06Rik |
RIKEN cDNA 2010203P06 gene |
6856 |
0.18 |
| chr14_57057660_57057892 | 0.26 |
Gja3 |
gap junction protein, alpha 3 |
53 |
0.97 |
| chr9_56994536_56995264 | 0.26 |
Ptpn9 |
protein tyrosine phosphatase, non-receptor type 9 |
23 |
0.97 |
| chr14_34560304_34560455 | 0.26 |
Ldb3 |
LIM domain binding 3 |
16615 |
0.11 |
| chr16_29977900_29978051 | 0.26 |
Gm1968 |
predicted gene 1968 |
1209 |
0.44 |
| chr2_32884809_32884960 | 0.26 |
Fam129b |
family with sequence similarity 129, member B |
8770 |
0.11 |
| chr11_62789333_62789872 | 0.26 |
Zfp286 |
zinc finger protein 286 |
140 |
0.92 |
| chr10_127516477_127517046 | 0.25 |
Ndufa4l2 |
Ndufa4, mitochondrial complex associated like 2 |
1794 |
0.18 |
| chr9_108480057_108480354 | 0.25 |
Lamb2 |
laminin, beta 2 |
324 |
0.71 |
| chr19_10304418_10305190 | 0.25 |
Dagla |
diacylglycerol lipase, alpha |
53 |
0.97 |
| chr8_69654078_69655164 | 0.25 |
Zfp964 |
zinc finger protein 964 |
129 |
0.95 |
| chr1_189615380_189615531 | 0.25 |
Gm38122 |
predicted gene, 38122 |
24779 |
0.18 |
| chr3_105018285_105018436 | 0.25 |
Gm40117 |
predicted gene, 40117 |
24328 |
0.13 |
| chr15_76206813_76207932 | 0.25 |
Plec |
plectin |
881 |
0.36 |
| chr8_122546722_122547355 | 0.24 |
Piezo1 |
piezo-type mechanosensitive ion channel component 1 |
4291 |
0.11 |
| chr16_92345192_92345711 | 0.24 |
Kcne1 |
potassium voltage-gated channel, Isk-related subfamily, member 1 |
13423 |
0.11 |
| chr8_71381974_71382671 | 0.24 |
Nr2f6 |
nuclear receptor subfamily 2, group F, member 6 |
362 |
0.74 |
| chr7_136406485_136406636 | 0.24 |
Gm36849 |
predicted gene, 36849 |
53196 |
0.14 |
| chr4_117985332_117985486 | 0.23 |
9530034E10Rik |
RIKEN cDNA 9530034E10 gene |
10373 |
0.15 |
| chr5_134742453_134742604 | 0.23 |
Gm30003 |
predicted gene, 30003 |
4514 |
0.16 |
| chr11_70608772_70609053 | 0.23 |
Mink1 |
misshapen-like kinase 1 (zebrafish) |
826 |
0.32 |
| chr11_59364469_59364771 | 0.23 |
Gm15755 |
predicted gene 15755 |
10841 |
0.12 |
| chr8_24438713_24439074 | 0.23 |
Tcim |
transcriptional and immune response regulator |
91 |
0.96 |
| chr15_6386264_6386948 | 0.23 |
Dab2 |
disabled 2, mitogen-responsive phosphoprotein |
8 |
0.98 |
| chr17_56672739_56673924 | 0.23 |
Ranbp3 |
RAN binding protein 3 |
27 |
0.96 |
| chr7_49527689_49528213 | 0.22 |
Nav2 |
neuron navigator 2 |
20241 |
0.23 |
| chr1_60652807_60652958 | 0.22 |
Gm23762 |
predicted gene, 23762 |
39079 |
0.1 |
| chr15_83724974_83725683 | 0.22 |
Scube1 |
signal peptide, CUB domain, EGF-like 1 |
307 |
0.92 |
| chr17_66448520_66450314 | 0.22 |
Mtcl1 |
microtubule crosslinking factor 1 |
333 |
0.57 |
| chr11_30648868_30649937 | 0.22 |
Acyp2 |
acylphosphatase 2, muscle type |
185 |
0.95 |
| chr1_93754379_93755416 | 0.22 |
Atg4b |
autophagy related 4B, cysteine peptidase |
8 |
0.5 |
| chr6_65670118_65670269 | 0.22 |
Ndnf |
neuron-derived neurotrophic factor |
1397 |
0.49 |
| chr9_44734596_44735576 | 0.22 |
Phldb1 |
pleckstrin homology like domain, family B, member 1 |
112 |
0.91 |
| chr14_47921837_47922130 | 0.21 |
4930447J18Rik |
RIKEN cDNA 4930447J18 gene |
23074 |
0.15 |
| chr9_60837244_60837770 | 0.21 |
Gm9869 |
predicted gene 9869 |
692 |
0.7 |
| chr7_143777485_143777636 | 0.21 |
Gm44998 |
predicted gene 44998 |
2447 |
0.15 |
| chr9_35331987_35332138 | 0.21 |
Gm33838 |
predicted gene, 33838 |
24463 |
0.11 |
| chr13_43864441_43864954 | 0.21 |
Gm33195 |
predicted gene, 33195 |
28 |
0.97 |
| chr4_129928556_129928707 | 0.21 |
Spocd1 |
SPOC domain containing 1 |
618 |
0.62 |
| chr10_25359547_25360523 | 0.21 |
Epb41l2 |
erythrocyte membrane protein band 4.1 like 2 |
179 |
0.95 |
| chr6_141249885_141250621 | 0.20 |
Gm28523 |
predicted gene 28523 |
244 |
0.55 |
| chr11_53904266_53904446 | 0.20 |
Gm12218 |
predicted gene 12218 |
1201 |
0.29 |
| chr2_145787427_145787839 | 0.20 |
Rin2 |
Ras and Rab interactor 2 |
1471 |
0.48 |
| chr9_41889978_41890642 | 0.20 |
Gm40513 |
predicted gene, 40513 |
294 |
0.89 |
| chr2_27212365_27212516 | 0.20 |
Sardh |
sarcosine dehydrogenase |
2503 |
0.21 |
| chr12_32378513_32379490 | 0.20 |
Ccdc71l |
coiled-coil domain containing 71 like |
297 |
0.93 |
| chr3_146117348_146117511 | 0.20 |
Mcoln3 |
mucolipin 3 |
21 |
0.96 |
| chr3_99183530_99183681 | 0.20 |
Gm18982 |
predicted gene, 18982 |
2324 |
0.2 |
| chr2_26491019_26491170 | 0.19 |
Notch1 |
notch 1 |
12728 |
0.1 |
| chr8_94377668_94378018 | 0.19 |
Gm15889 |
predicted gene 15889 |
29 |
0.95 |
| chr7_99626901_99627081 | 0.19 |
Tpbgl |
trophoblast glycoprotein-like |
112 |
0.9 |
| chr10_13473756_13474638 | 0.19 |
Phactr2 |
phosphatase and actin regulator 2 |
215 |
0.94 |
| chr5_112166735_112167033 | 0.19 |
1700016B01Rik |
RIKEN cDNA 1700016B01 gene |
10647 |
0.17 |
| chr1_88701358_88702305 | 0.19 |
Arl4c |
ADP-ribosylation factor-like 4C |
45 |
0.97 |
| chr9_70243232_70243383 | 0.19 |
Myo1e |
myosin IE |
35939 |
0.17 |
| chr4_149999267_149999418 | 0.19 |
H6pd |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
1923 |
0.28 |
| chr11_107547352_107548463 | 0.19 |
Helz |
helicase with zinc finger domain |
23 |
0.88 |
| chr19_5567497_5568487 | 0.19 |
Ap5b1 |
adaptor-related protein complex 5, beta 1 subunit |
33 |
0.93 |
| chr13_31798221_31798372 | 0.18 |
Gm11379 |
predicted gene 11379 |
757 |
0.63 |
| chr7_31115171_31115481 | 0.18 |
Hpn |
hepsin |
36 |
0.95 |
| chr8_120804879_120805047 | 0.18 |
Irf8 |
interferon regulatory factor 8 |
52754 |
0.1 |
| chr4_142077883_142078172 | 0.18 |
Tmem51os1 |
Tmem51 opposite strand 1 |
5945 |
0.15 |
| chr4_120144444_120144645 | 0.18 |
Edn2 |
endothelin 2 |
16662 |
0.22 |
| chr11_103244905_103245215 | 0.17 |
Map3k14 |
mitogen-activated protein kinase kinase kinase 14 |
2789 |
0.17 |
| chr7_66427385_66427727 | 0.17 |
Aldh1a3 |
aldehyde dehydrogenase family 1, subfamily A3 |
39 |
0.97 |
| chrY_90771840_90772811 | 0.17 |
Gm47283 |
predicted gene, 47283 |
12413 |
0.17 |
| chr17_31686321_31686544 | 0.17 |
Gm50222 |
predicted gene, 50222 |
2058 |
0.19 |
| chr5_99375821_99375972 | 0.17 |
Gm35394 |
predicted gene, 35394 |
101801 |
0.07 |
| chr5_106801577_106801728 | 0.17 |
Gm8365 |
predicted gene 8365 |
62888 |
0.09 |
| chr8_13695607_13695765 | 0.17 |
Cfap97d2 |
CFAP97 domain containing 2 |
10203 |
0.16 |
| chr17_71782235_71783145 | 0.17 |
Clip4 |
CAP-GLY domain containing linker protein family, member 4 |
73 |
0.97 |
| chr10_79866106_79866295 | 0.17 |
Plppr3 |
phospholipid phosphatase related 3 |
1081 |
0.19 |
| chr5_33695613_33695860 | 0.17 |
Gm42965 |
predicted gene 42965 |
12762 |
0.1 |
| chr11_75462938_75463374 | 0.17 |
Mir22 |
microRNA 22 |
560 |
0.47 |
| chr5_111532860_111533036 | 0.17 |
C130026L21Rik |
RIKEN cDNA C130026L21 gene |
48474 |
0.13 |
| chr19_5414445_5414596 | 0.17 |
Gm6293 |
predicted pseudogene 6293 |
1239 |
0.16 |
| chr13_106936192_106937354 | 0.17 |
Ipo11 |
importin 11 |
172 |
0.93 |
| chr15_76465093_76465561 | 0.17 |
Scx |
scleraxis |
7807 |
0.08 |
| chr10_42614130_42614512 | 0.16 |
Ostm1 |
osteopetrosis associated transmembrane protein 1 |
30483 |
0.16 |
| chr1_182090368_182090591 | 0.16 |
Gm37177 |
predicted gene, 37177 |
19076 |
0.14 |
| chr1_184731813_184732486 | 0.16 |
Hlx |
H2.0-like homeobox |
470 |
0.75 |
| chr4_133680182_133680333 | 0.16 |
Pigv |
phosphatidylinositol glycan anchor biosynthesis, class V |
7610 |
0.13 |
| chr1_33488501_33488652 | 0.16 |
Gm29228 |
predicted gene 29228 |
6475 |
0.23 |
| chr2_152734500_152735322 | 0.16 |
Id1 |
inhibitor of DNA binding 1, HLH protein |
1340 |
0.29 |
| chr6_72235086_72235414 | 0.16 |
Atoh8 |
atonal bHLH transcription factor 8 |
20 |
0.97 |
| chr1_31017458_31017792 | 0.16 |
4931428L18Rik |
RIKEN cDNA 4931428L18 gene |
17721 |
0.14 |
| chr11_86993066_86994378 | 0.16 |
Ypel2 |
yippee like 2 |
15 |
0.97 |
| chr7_6382694_6383817 | 0.16 |
Zfp28 |
zinc finger protein 28 |
40 |
0.94 |
| chr3_100462225_100462649 | 0.16 |
Gm43121 |
predicted gene 43121 |
24603 |
0.11 |
| chr11_97415706_97416357 | 0.15 |
Arhgap23 |
Rho GTPase activating protein 23 |
498 |
0.76 |
| chr10_62792164_62792669 | 0.15 |
Ccar1 |
cell division cycle and apoptosis regulator 1 |
130 |
0.93 |
| chr15_102105460_102106270 | 0.15 |
Tns2 |
tensin 2 |
753 |
0.51 |
| chr11_119807843_119807994 | 0.15 |
Rptor |
regulatory associated protein of MTOR, complex 1 |
3328 |
0.23 |
| chr19_43440308_43441144 | 0.15 |
Gm47936 |
predicted gene, 47936 |
266 |
0.5 |
| chr6_97311994_97312190 | 0.15 |
Frmd4b |
FERM domain containing 4B |
288 |
0.93 |
| chr18_61056422_61056649 | 0.15 |
Pdgfrb |
platelet derived growth factor receptor, beta polypeptide |
11335 |
0.13 |
| chr2_148044552_148044703 | 0.15 |
Foxa2 |
forkhead box A2 |
837 |
0.56 |
| chr9_107908926_107909153 | 0.15 |
Mst1r |
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
2132 |
0.13 |
| chr8_102786860_102787011 | 0.15 |
Cdh11 |
cadherin 11 |
1293 |
0.45 |
| chr2_91656850_91657112 | 0.15 |
Arhgap1 |
Rho GTPase activating protein 1 |
3773 |
0.13 |
| chr7_28277569_28278227 | 0.14 |
Eid2b |
EP300 interacting inhibitor of differentiation 2B |
159 |
0.89 |
| chr17_10093886_10094100 | 0.14 |
Qk |
quaking |
119581 |
0.06 |
| chr10_80656363_80657283 | 0.14 |
Btbd2 |
BTB (POZ) domain containing 2 |
248 |
0.82 |
| chr2_75997808_75997959 | 0.14 |
Pde11a |
phosphodiesterase 11A |
8160 |
0.17 |
| chr7_130865550_130866588 | 0.14 |
Plekha1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
159 |
0.63 |
| chr12_86974721_86975193 | 0.14 |
Zdhhc22 |
zinc finger, DHHC-type containing 22 |
8878 |
0.15 |
| chr18_76035512_76035730 | 0.14 |
1700003O11Rik |
RIKEN cDNA 1700003O11 gene |
19944 |
0.19 |
| chr11_106036361_106037705 | 0.14 |
Dcaf7 |
DDB1 and CUL4 associated factor 7 |
133 |
0.92 |
| chr7_133047426_133047577 | 0.14 |
Ctbp2 |
C-terminal binding protein 2 |
17385 |
0.17 |
| chr11_33871976_33872356 | 0.14 |
Kcnip1 |
Kv channel-interacting protein 1 |
28581 |
0.21 |
| chr9_48835918_48836556 | 0.13 |
Zbtb16 |
zinc finger and BTB domain containing 16 |
15 |
0.98 |
| chr15_103366694_103366859 | 0.13 |
Itga5 |
integrin alpha 5 (fibronectin receptor alpha) |
13 |
0.96 |
| chr15_103169326_103169551 | 0.13 |
Smug1 |
single-strand selective monofunctional uracil DNA glycosylase |
2346 |
0.18 |
| chr15_86202694_86203051 | 0.13 |
Gm22818 |
predicted gene, 22818 |
9785 |
0.16 |
| chr2_153256681_153256832 | 0.13 |
Pofut1 |
protein O-fucosyltransferase 1 |
726 |
0.59 |
| chr7_97737443_97738488 | 0.13 |
Aqp11 |
aquaporin 11 |
27 |
0.97 |
| chr17_47842318_47842529 | 0.13 |
Gm25201 |
predicted gene, 25201 |
1438 |
0.29 |
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.4 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.5 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.1 | 0.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.4 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.2 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) |
| 0.1 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.3 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.5 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.1 | 0.2 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.2 | GO:2000611 | positive regulation of thyroid hormone generation(GO:2000611) |
| 0.1 | 0.2 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 0.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.0 | 0.3 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.5 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.3 | GO:0034651 | cortisol biosynthetic process(GO:0034651) |
| 0.0 | 0.2 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.6 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.0 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.1 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0051138 | regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.0 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.2 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.0 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.2 | GO:0045821 | positive regulation of glycolytic process(GO:0045821) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.0 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.0 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.0 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0071225 | cellular response to muramyl dipeptide(GO:0071225) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.0 | 0.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.2 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.6 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0001031 | RNA polymerase III type 1 promoter DNA binding(GO:0001030) RNA polymerase III type 2 promoter DNA binding(GO:0001031) RNA polymerase III type 3 promoter DNA binding(GO:0001032) 5S rDNA binding(GO:0080084) |
| 0.0 | 1.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.0 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |